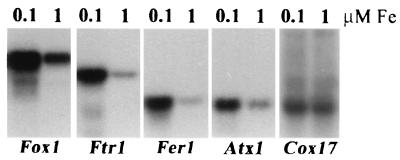
FIG. 1.
Increased accumulation of RNAs encoding iron metabolism components in iron-deficient Chlamydomonas cells. C. reinhardtii cells from a late-log culture in copper-free TAP medium were harvested and resuspended in 90 ml of −Cu TAP with 0.1 μM iron chelate. One milliliter was used to inoculate 100-ml cultures of +Cu TAP with either 0.1 μM iron chelate (cells severely chlorotic) or 1 μM iron chelate (chlorophyll content relatively unaffected compared to that of iron-replete cells). Cultures were grown to late log phase and transferred, and this process was repeated twice more to adapt cells to 1 or 0.1 μM iron chelate. Total RNA was prepared from late-log-phase cultures after three rounds and analyzed by hybridization with gene-specific probes as indicated. For quantitation, the signals were normalized to total RNA loaded. Specifically, the relative intensities (10−5) after object average background correction were as follows: Fox1, 25.2 and 8.7; Ftr1, 5.2 and 1.2; Fer1, 3.7 and 0.9; Atx1, 1.2 and 0.5; Cox17, 0.05 and 0.05. For RbcS2, the signal was actually decreased in 0.1 μM iron samples. The relative intensities (10−5) were 20.6 and 44.8. Transcript sizes were as follows: Fox1, 5.2 kb; Ftr1, 2.8 kb; Fer1, 1.3 kb; and Atx1, 1.0 kb. They were estimated from a standard curve of the relative mobility of each marker (Gibco BRL; 0.24- to 9.5-kb RNA marker) versus log10 of its size in bases.