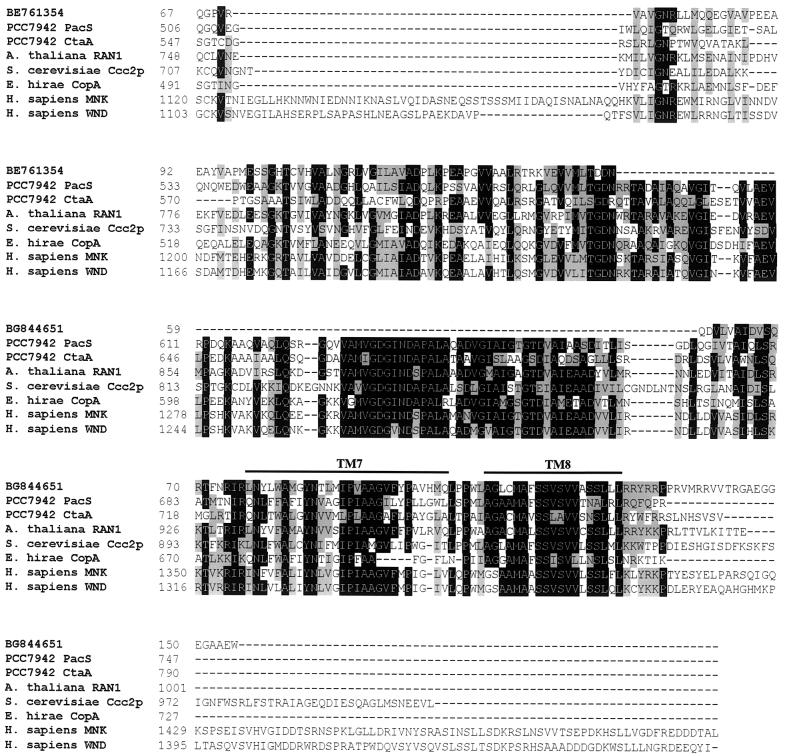
FIG. 11.
Sequence comparison between C. reinhardtii ESTs and copper ATPases. The predicted amino acid sequences encoded by C. reinhardtii ESTs (GenBank accession no. BE761354 and BG844651) were aligned with the relevant regions of copper-transporting ATPases from other organisms. The alignment was generated by using the ClustalW algorithm and BioEdit software (34). The numbers indicate the positions of the amino acids in each sequence. The first sequence in the alignment represents the amino acids encoded by the two C. reinhardtii ESTs, which align with different regions of the copper ATPases but are derived from a single gene. The alignment of the ESTs was based on BLAST output. Residues that are similar or identical in a majority (five) of sequences are shaded gray and black, respectively. A line above the alignment indicates the transmembrane regions. GenBank accession numbers: Synechococcus PacS, P37279; Synechococcus CtaA, AAB82020; A. thaliana RAN1, AF082565; S. cerevisiae Ccc2p, L36317; E. hirae CopA, L13292; H. sapiens MNK, NM_000052; H. sapiens WND, NM_000053.