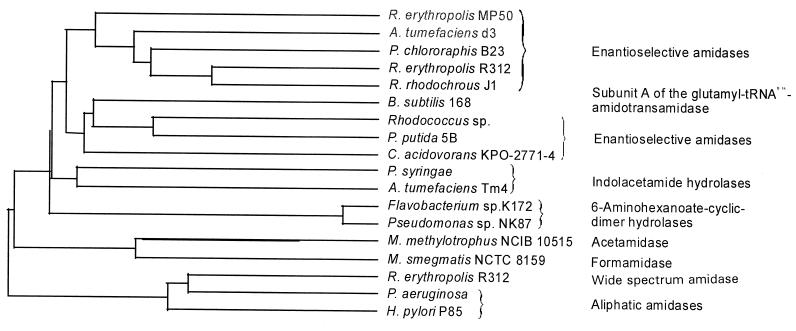
FIG. 4.
Dendrogram resulting from pairwise alignments of amino acid sequences by using the program CLUSTAL. Agrobacterium tumefaciens d3 (AF315580 [60]); Agrobacterium tumefaciens (P03868 [30]); Bacillus subtilis 168 (O06491 [14]); Comamonas acidovorans KPO-2771-4 (20); Flavobacterium sp. strain K172 (P13397 [61]); Helicobacter pylori 85P (CAA72932 [53]); Methylophilus methylotrophus (Q50228 [67]); Mycobacterium smegmatis NCTC 8159 (Q07838 [42]), Pseudomonas aeruginosa (P11436 [2]), Pseudomonas chlororaphis B23 (P27765 [49]); Pseudomonas putida 5B (O69768 [67]); Pseudomonas sp. strain NK87 (P13398 [61]); Pseudomonas syringae EW2009 (P06618 [69]); Rhodococcus rhodochrous J1 (S38270 [32]); Rhodococcus sp. (A41326 [45]); Rhodococcus sp. strain R312 (enantioselective amidase) (P22984 [44]); and Rhodococcus sp. strain R312 (“wide-spectrum” amidase) (Q01360 [56]).