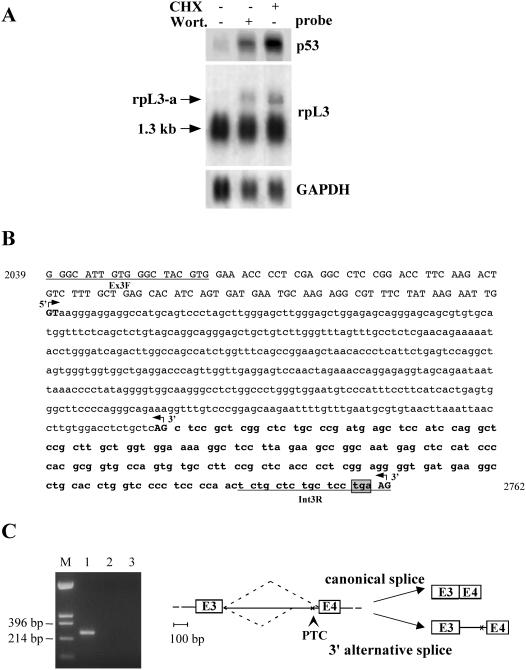
Figure 1.
Identification of NMD sensitive isoforms of the human rpL3. (A) Analysis of p53 and rpL3 expression in Calu-6 cells after incubation with CHX (4 h) or wortmannin (Wort.) (2 h). Total RNA purified from treated or untreated cells was hybridized with the cDNA specific probes as indicated. The size of canonical mRNA is indicated; rpL3-a indicates the newly identified isoform. The quantity of RNA was normalized using a GAPDH-specific probe. A representative result of three independent experiments is shown. (B) Nucleotide sequence of rpL3 intron 3 with 5′ flanking exonic sequences. Lower case letters refer to intronic nucleotides; upper case letters refer to exonic nucleotides. The intronic sequence retained by alternative splicing is indicated in bold. The primers used for RT–PCR are underlined. Canonical and alternative splice sites (GT/AG), as determined by sequence analysis of RT–PCR products, are shown in bold upper case letters and indicated by 5′ or 3′ arrows. The ORF of exonic and retained intronic sequences is shown. Nucleotides encoding the PTC are boxed and shaded in gray. The nucleotide positions according to the corresponding NCBI/GenBank sequences (accession no.: AL022326) is indicated. (C) Left panel RT–PCR assays of RNA from Calu-6 cells. Amplifications were performed with the following primers: lane 1, Ex3F/Int3R (rpL3-specific); lane 2, reaction was carried out as in lane 1 but omitting the RT step. As negative control, amplification was performed without cDNA (lane 3). The base pairs of HinfII-digested pUC18 (lane M) are indicated on the left. Right panel, schematic drawing of alternative splicing of human rpL3; intron 3 and flanking exons 3 and 4 (E3, E4) are shown.