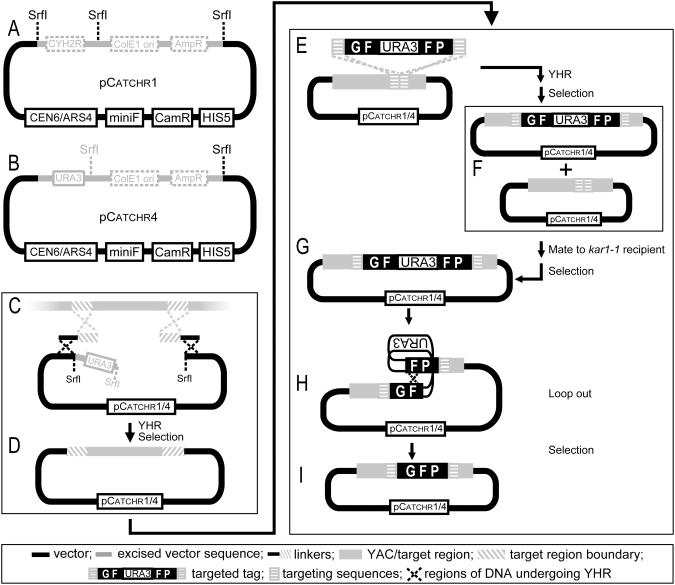
Figure 2.
Overview of the Gene Catchr method. (A–D) Cloning steps; (E–I) tagging steps. Schematic representations of the shuttle vectors (A) pCatchr1 and (B) pCatchr4. pCatchr1/4 has two origins of replication for E.coli: miniF for low-copy plasmid maintenance, and ColE1 for high-copy plasmid maintenance. The ColE1 origin allows for efficient amplification of pCatchr1/4, but is removed by SrfI digestion once a large insert is subcloned, to prevent rearrangements often associated with large high-copy plasmids (33,63,64). Linearization of pCatchr1/4 with SrfI produces a vector with free ends, and a URA3 overhanging fragment in the case of pCatchr4 only [(C) shown in gray]. Subsequent selection against the URA3 marker ensures that pCatchr4 plasmids incorrectly formed by recircularization are eliminated. (C) Linkers contain 450–700 bp of homology to the target region boundaries and 40 bp of homology to the free vector ends. Yeast containing the appropriate YAC are transformed with SrfI-linearized pCatchr1/4 and linkers. (D) Correct plasmids are formed by YHR, and contain the target region cloned into pCatchr1/4. (E–I) Summary of the Gene Catchr tagging method. (E) The GF-URA3-FP targeted-tag contains the GFP coding sequence with 288 bp direct repeats (‘F’) encoding parts of gfp exons 2 and 3, interrupted by the URA3 marker, and flanked by targeting sequences (40–50 bp) homologous to the insertion site in the cloned gene. (E and F) GF-URA3-FP is inserted into the cloned worm gene in pCatchr1/4 by YHR. (F) After selection, vectors containing both tagged and untagged genes are present in the resulting yeast colonies. (F and G) Exceptional cytoduction (mating to a kar1-1 recipient strain and subsequent selection steps) is used to isolate yeast cells containing only the tagged plasmids. (H and I) Direct repeats (‘F’) in the gfp sequence allow for the excision of the URA3 marker, thereby reconstituting the GFP tag.