Figure 2.
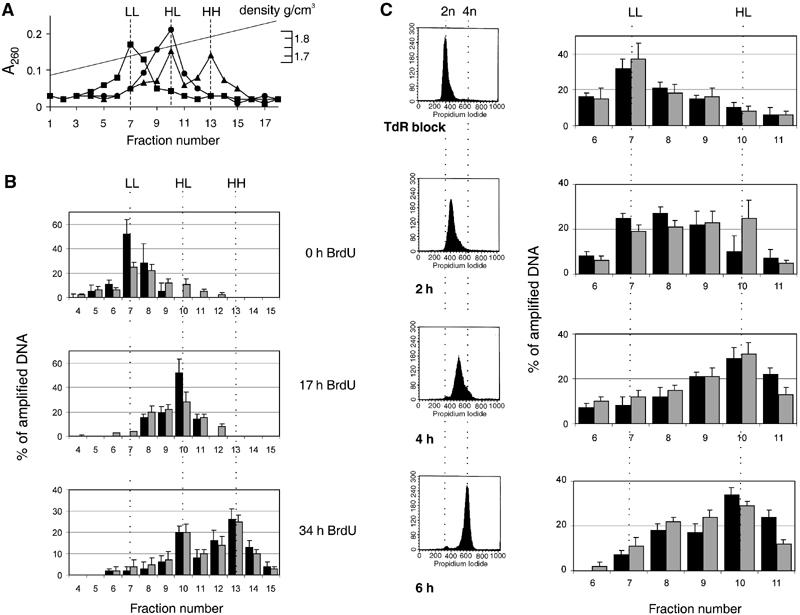
pEPI-1 replicates in a once-per-cell-cycle manner synchronously with cellular DNA and early in the S phase. Asynchronously growing CHO cells were labeled with 30 μg/ml BrdU for 0, 17 or 34 h. After shearing and digestion, total DNA was separated by CsCl gradient centrifugation. (A) The distribution of total DNA was determined from A260 measurements. Squares, 0 h; circles, 17 h; triangles, 34 h. Three peaks of A260-absorbing material were seen at densities of 1.7, 1.75 and 1.8 g/cm3 corresponding to light–light (LL), heavy–light (HL) and heavy–heavy (HH) DNA strands. (B) Samples were dialyzed against TE, precipitated and further analyzed by quantitative PCR using primer pairs SV40 for the plasmid pEPI-1 and DHFRex1 for genomic CHO DNA. The distribution of pEPI-1 (gray bars) or cellular DNA (black bars) is presented as a percentage of total amplified DNA. (C) CHO cells were synchronized by the excess-thymidine procedure at the G1-to-S-phase transition and then released into thymidine-free medium containing BrdU. Left, FACS analysis of isolated nuclei. Right, CsCl density gradient centrifugation followed by quantitative PCR to determine the distribution of cellular DNA (black bars) and plasmid DNA (gray bars).
