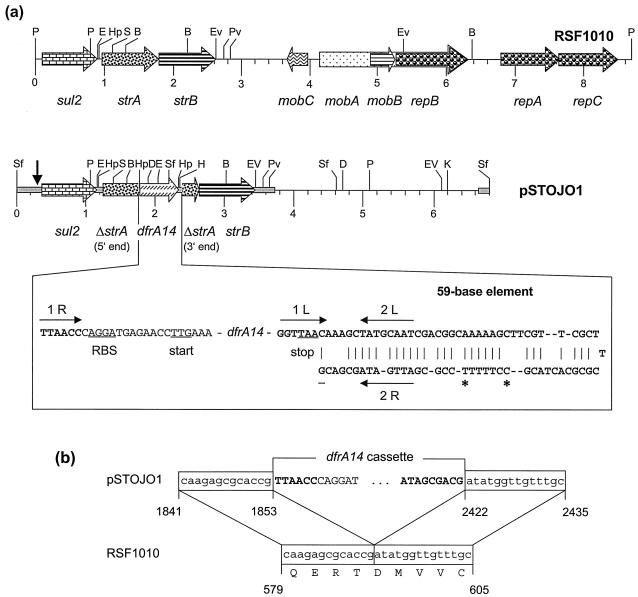
FIG. 1.
(a) Restriction map and structural organization of plasmids RSF1010 (8) and pSTOJO1. Restriction endonucleases: B, BclI; D, DraI; E, EcoRI; EV, EcoRV; H, HindIII; Hp, HpaI; K, KpnI; P, PstI; Pv, PvuII; S, SacI; and Sf, SfuI. A distance scale in kilobases is presented below the map. The reading frames for the genes sul2, ΔstrA, strA, dfrA14, strB, mobA-C, and repA-C are shown as arrows, with the direction of transcription indicated by the arrowhead. The gray bar in pSTOJO1 indicates the sequenced part. The vertical arrow at the left end of the map of pSTOJO indicates the beginning of the RSF1010-related part of the pSTOJO1 sequence. Essential parts of the dfrA14 cassette are shown in more detail below the map of pSTOJO1. The ribosome binding site (RBS) and translational start and stop codons are underlined. In the 59-be, the putative IntI1 integrase binding domains 1L, 2L, 2R, and 1R (11) are indicated by arrows. The entire 59-be of the cassette is shown in boldface. The C marked with an asterisk is missing in the 59-be of the dfrA14 cassette of plasmid pUK1329 from E. coli (Z50805), and the T marked with an asterisk is missing in that of plasmid pHCM1 from S. enterica serovar Typhi (AL513383). (b) Comparison of the integration site of the dfrA14 cassette within strA in pSTOJO1 and the corresponding strA sequence of RSF1010 (8). The numbering refers to the positions in the database entries for pSTOJO1 (AJ313522) and RSF1010 (M28829).