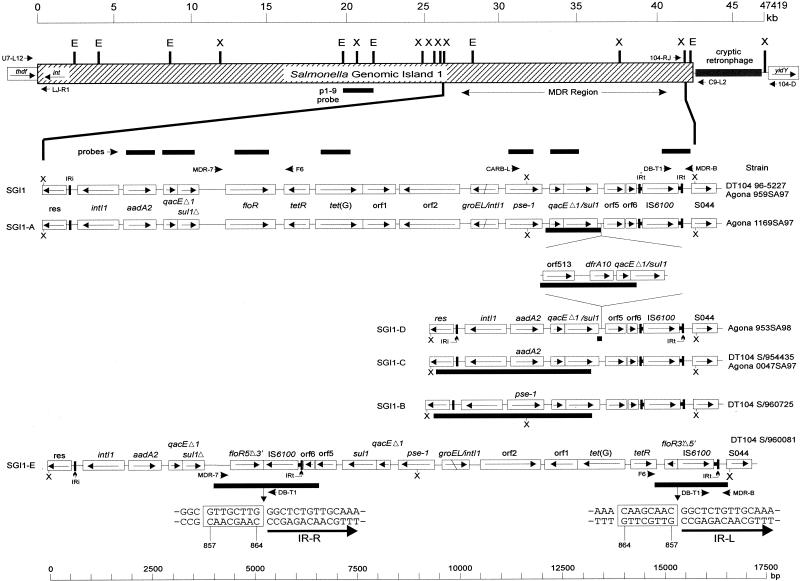
FIG. 1.
Genetic organization of the MDR regions of the various strains in this study based on Southern hybridization data, PCR mapping, and sequence analysis. A schematic of SGI1 is shown at the top, with the approximate locations of the primers used to detect the left and right junction regions indicated. The cryptic retronphage region is absent in Agona strains. Direction of transcription of genes is indicated by arrows. Locations of PCR products used as probes are indicated (thin black bars), as are the locations of some primers used in mapping. IRi and and IRt are 25-bp imperfect inverted repeats defining the ends of class 1 integrons. Regions sequenced are indicated under the various MDR regions by a thick black bar. The nucleotide sequence of the insertion point of the IS6100 element in the floR gene of S/960081 is shown; the 8-bp region duplicated upon insertion is boxed (coordinates are for the floR gene only). IR-L and IR-R refer to the inverted repeats that define one end of IS6100. The sequence of the orf513/dfrA10 region from Agona 1169SA97 has been assigned accession no. AY049746.