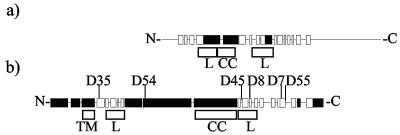
FIG. 1.
Representations of MexA and HlyD secondary structures. Black boxes, alpha helices; white boxes, beta strands. (a) MexA (383 amino acids) secondary structure taken from Protein Data Bank entry 1T5E. Only residues 29 to 259 are shown in the structure. (b) HlyD (478 amino acids) secondary structure prediction from the SAM-T02 server based upon 536 MFP sequences (http://www.cse.ucsc.edu/research/compbio/HMM-apps/T02-query.html). The positions of the mutations identified in HlyD are as indicated, together with the positions of the lipoyl motifs (boxed L), TMD (boxed TM), and coiled-coil domain (boxed CC). The precise amino acid changes in the different mutants are shown in Table 1. The first 40 residues forming an amphiphilic helix are essential for secretion of HlyA (40) being involved in the initial interaction with HlyA and triggering of TolC recruitment to the translocator (3, 49).