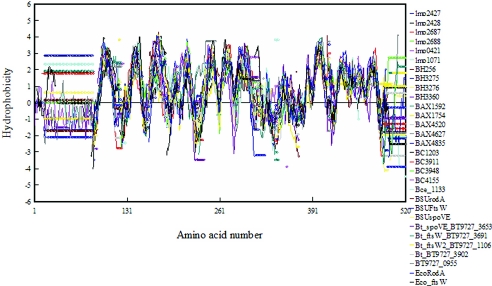
FIG. 8.
Multiple hydropathy alignment of RodA-FtsW-like proteins from L. monocytogenes and related species. Proteins belonging to the RodA-FtsW family were identified from the genomes of L. monocytogenes strain EGDe (Lmo), Bacillus subtilis 168 (BSU), Bacillus cereus ATCC10987 (BC), Bacillus anthracis Sterne (BAX), Bacillus thuringiensis serovar konkukian strain 97-27 (BT), Bacillus halodurans C-125 (BH), and Escherichia coli MG1655 (ECO). Multiple alignment was performed using CLUSTAL W (57) using a Blosum scoring matrix, and multiple hydrophobicity plots based on the alignment were generated based on the methods of Hopp and Woods (29, 37) as implemented in the DNAman package. The plot shown was developed using a window size of eight amino acid residues. Each colored plot and the corresponding protein are shown to the right of the plot. Circles in the plot indicate gaps in the alignment or start-stop positions.