Abstract
Klebsiella pneumoniae KOL, a clinical strain resistant to various β-lactams, was isolated from the stools of a patient from Greece. This strain harbored a new pI 9.1 plasmid-mediated AmpC β-lactamase with unusually high levels of hydrolytic activity for cefoxitin and cefotetan that we named MOX-2. Sequencing of blaMOX-2 revealed 93.2, 92.9, 92.7, and 73.1% identities with the deduced amino acid sequences of CMY-8, MOX-1, CMY-1, and the AmpC β-lactamase of Aeromonas sobria, respectively.
Plasmid-encoded AmpC-type β-lactamases have been described in clinical strains of various Enterobacteria. For some of them, the amino acid and nucleotide sequences of the corresponding genes are very similar to those of the chromosome-encoded AmpC β-lactamases of Enterobacter cloacae (ACT-1 and MIR-1), Citrobacter freundii (CMY-2, CMY-4, CMY-5, and LAT-1), Morganella morganii (DHA-1 and DHA-2), and Hafnia alvei (ACC-1) (15). The phylogeny of MOX-1, CMY-1, and FOX-type enzymes (5, 7, 12) is unclear as they show lower sequence similarities (≤77%) to those of the chromosomally encoded AmpC β-lactamases of Aeromonas sobria, Pseudomonas aeruginosa, and Serratia marcescens (15, 18).
A strain of Klebsiella pneumoniae (KOL) resistant to various β-lactam antibiotics, including cephamycins, was isolated in 1997 at Lariboisière Hospital, Paris, France, from the stools of a patient transferred from an intensive care unit in Athens, Greece, for treatment of a carotid cavernous fistula after a road accident (S. Boyer, L. Raskine, B. Hanau, A. Philippon, M. J. Sanson-Le-Pors, and G. Arlet, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C7, p. 70, 1998). He had previously received ampicillin-sulbactam and pefloxacin for chronic osteomyelitis and pulmonary infection. Our results showed that this strain harbored a new plasmid-encoded cephamycin-hydrolyzing β-lactamase belonging to the Aeromonas group identified by Philippon et al. (15) and for which we propose the designation MOX-2.
MICs were determined by using the agar dilution method recommended by the National Committee for Clinical Laboratory Standards (14). K. pneumoniae KOL was highly resistant to β-lactams (including penicillins, broad-spectrum cephalosporins, cephamycins, and aztreonam) except imipenem (Table 1). It was also resistant to aminoglycosides, trimethoprim, tetracycline, fluoroquinolones, and chloramphenicol (data not shown).
TABLE 1.
MICs of β-lactams for K. pneumoniae KOL; its transconjugant; the clones producing TEM-1, SHV-5, and MOX-2; E. coli 53-2; and E. coli JM101a
| Strain | Plasmid | bla | MIC (μg/ml) of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX | FOX | CTT | CAZ | CAZ-CLA | CTX | MOX | FEP | IMP | |||
| K. pneumoniae KOL | pKOL | MOX-2 | 1,024 | 1,024 | 64 | 256 | 8 | 8 | 4 | 4 | 0.125 |
| SHV-5 | |||||||||||
| TEM-1 | |||||||||||
| SHV-1 | |||||||||||
| E. coli 53-2 | 4 | 1 | 0.125 | 0.12 | 0.12 | <0.06 | |||||
| E. coli 53-2 TcKOL | pKOL | MOX-2 | 512 | 128 | 4 | 4 | 0.5 | 1 | 0.25 | 0.25 | 0.06 |
| SHV-5 | |||||||||||
| TEM-1 | |||||||||||
| E. coli JM101 | 4 | 4 | 0.125 | 0.25 | 0.25 | 0.06 | 0.25 | 0.06 | 0.03 | ||
| pLRB01 | MOX-2 | 64 | >1,024 | 512 | 32 | 32 | 128 | 32 | 0.25 | 0.25 | |
| pLRB02 | SHV-5 | 256 | 16 | 1 | 1,024 | 1 | 64 | 2 | 16 | 0.125 | |
| pLRB03 | TEM-1 | >1,024 | 4 | 0.125 | 0.5 | 0.5 | 0.06 | 0.125 | 0.5 | 0.125 | |
Abbreviations: bla, β-lactamase; AMX, amoxicillin; FOX, cefoxitin; CTT, cefotetan; CAZ, ceftazidime; CTX, cefotaxime; MOX, moxalactam; FEP, cefepime; IMP, imipenem; CLA, clavulanic acid (2 μg/ml).
An Escherichia coli J53-2 Rifr transconjugant (TcKOL) was selected on Mueller-Hinton plates supplemented with rifampin (250 μg/ml) and ticarcillin (100 μg/ml) or cefotaxime (1 μg/ml). TcKOL showed a β-lactam resistance phenotype similar to that of the parental strain, although the level of resistance was lower, suggesting the presence of an additional resistance mechanism in the KOL strain.
Plasmid DNA was extracted from the transconjugant according to the method of Kado and Liu for large plasmids (8). Its analysis revealed one plasmid of about 130 kb (data not shown).
Analytical isoelectric focusing was performed on a polyacrylamide gel with sonicated crude cell extracts as described previously (11). Three bands of β-lactamase activity with pIs of 9.1, 5.4, and 8.2 were detected in K. pneumoniae KOL and its transconjugant. A fourth band (pI 7.6) was present only in K. pneumoniae KOL (data not shown).
The β-lactamase with a pI of 9.1 from the transconjugant was characterized after purification as previously described (4). The kinetic constants, kcat and Km, for substrates were determined by computerized microacidimetric assay at pH 7.0 and 37°C in 0.1 M NaCl as described by Labia et al. (9). One unit of β-lactamase activity was defined as the amount of enzyme required to hydrolyze 1 μmol of benzylpenicillin per min at pH 7.0 and 37°C.
On the basis of the kcat values, MOX-2 hydrolyzed cefazolin 156 times faster than benzylpenicillin. Cefoxitin and cefotetan were hydrolyzed 7 and 3 times faster, respectively, than benzylpenicillin. These latter rates of hydrolysis were unusually high. The kcat values were about 100 times higher than the values generally reported for class C β-lactamases (13), but kcat/Km values were close to normal values, suggesting ready deacylation for these antibiotics. In contrast, the kcat of moxalactam remained low (kcat = 0.04 s−1), which is probably related to the carboxylate of the 7-α side chain of this molecule. In terms of Km, MOX-2 had a higher affinity for cefoxitin than for cephalothin or benzylpenicillin. The Ki values of MOX-2 for cefotaxime, ceftazidime, and aztreonam were lower than those usually observed, about 1/10 of those reported for chromosome-encoded enzymes (Table 2).
TABLE 2.
Kinetic parameters of purified MOX-2 β-lactamase
| Substrate | kcat (s−1) | Km (μM)a | kcat/Km (s−1 μM−1) |
|---|---|---|---|
| Benzylpenicillin | 5 | 9.7 | 0.51 |
| Cephalothin | 250 | 78 | 3.20 |
| Cephaloridine | 180 | 890 | 0.20 |
| Cefazolin | 800 | 712 | 1.12 |
| Cefotaxime | 0.05 | 0.9 | |
| Ceftazidime | 0.055 | 0.01 | |
| Cefepime | 1 | 20 | 0.05 |
| Aztreonam | <0.01 | ||
| Cefoxitin | 35 | 300 | 0.12 |
| Cefotetan | 15 | 42 | 0.35 |
| Moxalactam | 0.04 | 0.30 |
Cefotaxime, ceftazidime, aztreonam, and moxalactam had Ki values of 0.06, 4.5, 0.05, and 0.13 μM, respectively. Km values were not calculated for these substrates.
Fragments of the gene encoding the putative class C β-lactamase of K. pneumoniae KOL and of its transconjugant were amplified with degenerate oligonucleotide primers ampC A1, ampC A2, ampC B1, and ampC B2 as previously described (12). We amplified the putative TEM and SHV genes from K. pneumoniae KOL and its transconjugant with the primers OT3, OT4, OS5, and OS6, respectively, as previously described (2, 3).
Total DNA was extracted from K. pneumoniae KOL and its transconjugant as previously described (6) and partially or totally restricted with PstI and EcoRI plus HindIII. The resulting fragments were ligated to the vector pBK-CMV (Stratagene, La Jolla, Calif.) and digested with PstI or EcoRI plus HindIII. Recombinant plasmids were introduced into E. coli JM101 by the CaCl2 transformation method (16). Transformants with three different β-lactam susceptibility patterns (Table 1) were selected on Mueller-Hinton agar supplemented with kanamycin (50 μg/ml) and ampicillin (50 μg/ml). The sizes of the inserts of the recombinant plasmids, named pLRB01, pLRB02, and pLRB03, were estimated to be 6,170, 2,600, and 3,000 bp, respectively, by restriction enzyme digestion and electrophoresis on 1% agarose gels. The MICs for the clones were higher than those for the transconjugant, probably because there were fewer copies of the plasmid in TcKOL than of the multicopy plasmid vector pBK-CMV. The phenotype of the clone harboring pLRB01 was similar to that of strains overproducing AmpC cephalosporinase. DNA amplification showed that it produced the MOX-2 cephalosporinase with a pI of 9.1.
DNA was sequenced as described by Sanger et al. (17) with PCR primers or T3 and T7 universal sequencing primers, fluorescent dye-labeled dideoxynucleotides, Taq polymerase, and an ABI 373A DNA sequencer (Applied Biosytems, Foster City, Calif.).
The BLASTN program (1) at the National Center for Biotechnology Information was used for database searches. The ClustalW program (www.infobiogen.fr) was used to align multiple-protein sequences. Open reading frames (ORFs) were identified with the ORF Finder program (www.pasteur.fr).
DNA sequencing of the PCR products of the clones harboring pLRB02 and pLRB03 revealed that these clones produced an SHV-5-type β-lactamase (pI 8.2) and a TEM-1 β-lactamase (pI 5.4), respectively.
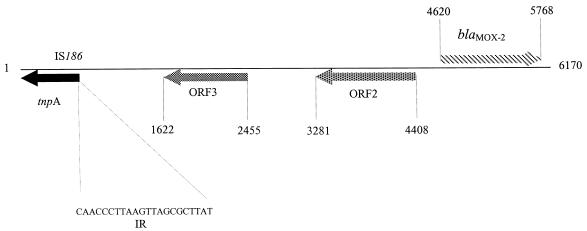
The 6,170-bp DNA insert of pLRB01 was sequenced, and four ORFs were identified (Fig. 1). An ORF of 1,149 bp (nucleotides [nt] 4620 to 5768) encoded a putative protein of 382 amino acids, with an estimated molecular mass of 38.5 kDa, preceded by putative Shine-Dalgarno and promoter consensus sequences [TTGGCG(N)16TACTTG]. The product of this ORF was most similar to the plasmid-encoded β-lactamases CMY-8 (19) (93.2% sequence identity), MOX-1 (7) (92.9% sequence identity), and CMY-1 (5) (92.7% sequence identity), followed by the chromosome-encoded AmpC β-lactamase of A. sobria (73.1% sequence identity). It showed a lower level of similarity to the AmpC β-lactamases of E. cloacae, C. freundii, M. morganii, H. alvei, E. coli (43 to 47% sequence identity), and P. aeruginosa (54% sequence identity). We therefore identified this ORF as blaMOX-2. There were several conserved serine β-lactamase motifs: the SXSK motif from the active site, the typical class C motif YXN, and the KTG domain. Alignment with the amino acid sequences of MOX-1, CMY-1, and CMY-8 showed two nonconservative substitutions in the region close to the active site: Ser-74→Arg and Thr-92→Pro (Fig. 2). Usually, the residue following element one, SXXK (residues 88 and 91) in class C β-lactamases, is a threonine. Element one is located at the beginning of the highly conserved helix H2. Thus, the presence of a proline in this position distorts this helix and, consequently, the active site. Analysis of the nucleotide sequence upstream from the start codon showed that no ampR gene was present.
FIG. 1.
Genetic organization of the genes identified on the 6,170-bp DNA insert of pLRB01. The directions of gene transcription are indicated by arrows. IR, inverted repeat.
FIG. 2.
Multiple-sequence alignment of the deduced amino acid sequence of the MOX-2 β-lactamase with those of the MOX-1, CMY-1, and CMY-8 β-lactamases. Dashes indicate identical amino acids.
The other ORFs were on the opposite DNA strand. ORF2 is located 212 bp upstream from blaMOX-2 and encodes a putative 375-amino-acid protein 40% identical to the transposase of the IS1358 from Vibrio anguillarum (10) (accession no. VAU93590). ORF3 is located 826 nt upstream from ORF2 and encodes a putative protein of 275 amino acids displaying no significant similarity to any protein in the database. A 612-bp ORF (nt 1 to 612) lacking 5′ sequences was identified 1,010 bp upstream from ORF3. It encodes a 204-amino-acid product 100% identical to the putative transposase of IS186 of E. coli K12 (accession no. X03123). A 22-bp inverted repeat was identified 32 nt downstream (nt 645 to 666) from this ORF.
We analyzed the regions flanking blaMOX-2 and did not find the characteristic structures of an integron or sequences similar to those flanking blaCMY-8 (19), blaMOX-1 (7), and blaCMY-1 (5). The means by which the blaMOX-2 gene was inserted were not clear.
We did not determine the exact phylogenetic origin of blaMOX-2. Its G+C content of 63.4% and its observed 73.1% identity with the A. sobria cephalosporinase suggest that the parental strain may have been a bacterium of the Aeromonas genus (G+C content of 57 to 63%).
Nucleotide sequence accession number.
The EMBL accession number for the nucleotide sequence reported in this paper is AJ276453.
Acknowledgments
We thank Alain Philippon for expert scientific advice and Fabienne Meunier for excellent technical assistance.
This work was partly supported by a grant from the Ministère de l'Education Nationale, de la Recherche et de la Technologie (Réseau β-lactamase), Paris, France, and by a grant from Bristol Laboratories.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arlet, G., G. Brami, D. Decré, A. Flippo, O. Gaillot, P. H. Lagrange, and A. Philippon. 1995. Molecular characterisation by PCR-fragment length polymorphism of TEM β-lactamase. FEMS Microbiol. Lett. 134:203-208. [DOI] [PubMed] [Google Scholar]
- 3.Arlet, G., M. Rouveau, and A. Philippon. 1997. Substitution of alanine at position 179 in the SHV-6 extended spectrum β-lactamase. FEMS Microbiol. Lett. 152:163-167. [DOI] [PubMed] [Google Scholar]
- 4.Barnaud, G., R. Labia, L. Raskine, M. J. Sanson-Le Pors, A. Philippon, and G. Arlet. 2001. Extension of resistance to cefepime and cefpirome associated to a six amino acid deletion in the H-10 helix of the cephalosporinase of an Enterobacter cloacae clinical isolate. FEMS Microbiol. Lett. 195:185-190. [DOI] [PubMed] [Google Scholar]
- 5.Bauernfeind, A., I. Stemplinger, R. Jungwirth, R. Wilhelm, and Y. Chong. 1996. Comparative characterization of the cephamycinase blaCMY-1 gene and its relationship with other β-lactamase genes. Antimicrob. Agents Chemother. 40:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimont, F., and P. A. D. Grimont. 1986. Ribosomal nucleic acid gene restriction as potential taxonomic tools. Ann. Inst. Pasteur Microbiol. 137B:165-175. [DOI] [PubMed] [Google Scholar]
- 7.Horii, T., Y. Arakawa, M. Ohta, T. Sugiyama, R. Wacharotayankun, H. Ito, and N. Kato. 1994. Characterization of a plasmid-borne and constitutively expressed blaMOX-1 gene encoding AmpC-type β-lactamase. Gene 139:93-98. [DOI] [PubMed] [Google Scholar]
- 8.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labia, R., J. Andrillon, and F. Le Goffic. 1973. Computerized microacidimetric determination of a β-lactamase Michaelis Menten constants. FEBS Lett. 33:42-44. [DOI] [PubMed] [Google Scholar]
- 10.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchese, A., G. Arlet, G. C. Shito, P. H. Lagrange, and A. Philippon. 1996. Detection of SHV-5 extended-spectrum beta-lactamase in Klebsiella pneumoniae strains isolated in Italy. Eur. J. Microbiol. Infect. Dis. 15:245-248. [DOI] [PubMed] [Google Scholar]
- 12.Marchese, A., G. Arlet, G. C. Shito, P. H. Lagrange, and A. Philippon. 1998. Characterization of FOX-3, an AmpC-type plasmid-mediated β-lactamase from an Italian isolate of Klebsiella oxytoca. Antimicrob. Agents Chemother. 42:464-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matagne, A., A. Dubus, M. Galleni, and J. M. Frère. 1999. The β-lactamase cycle: a tale of selective pressure and bacterial ingenuity. Nat. Prod. Rep. 16:1-19. [DOI] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 15.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh, T. R., L. Hall, A. P. MacGowan, and P. M. Bennett. 1995. Sequence analysis of two chromosomally mediated inducible β-lactamases from Aeromonas sobria, strain 163a, one a class D penicillinase, the other an AmpC cephalosporinase. J. Antimicrob. Chemother. 36:41-52. [DOI] [PubMed] [Google Scholar]
- 19.Yan, J. J., S. M. Wu, S. H. Tsai, J. J. Wu, and I. J. Su. 2000. Prevalence of SHV-12 among clinical isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamases and identification of a novel AmpC enzyme (CMY-8) in Southern Taiwan. Antimicrob. Agents Chemother. 44:1348-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]