FIG. 4.
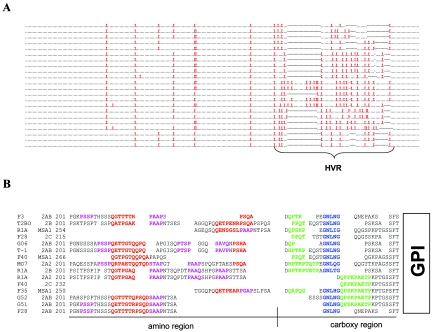
The HVR of MSA-2a/b contains a series of three, proline-rich, repeating motifs. (A) Amino acid alignment of all 22 MSA-2a/b proteins examined. A period replaces all nonproline residues, a red “X” replaces all proline residues, and dashes indicate gaps in sequences. (B) Manual amino acid alignment of 10 representative MSA-2a/b HVRs along with the 3′ region upstream of the GPI anchor signal sequence of F35, F40, and R1A MSA-1 and F40 and F28 MSA-2c. Alignments are based on both amino acid and nucleotide sequences. Motifs are highlighted by color. Sequences of the first motif are red, sequences of the second motif are purple, sequences of the third motif are green, and the common GNLNG sequence is blue. Residues that are not assigned to a motif are in black letters. Blank spaces indicate gaps. A black bisected underline splits the HVR into amino and carboxy regions as labeled.