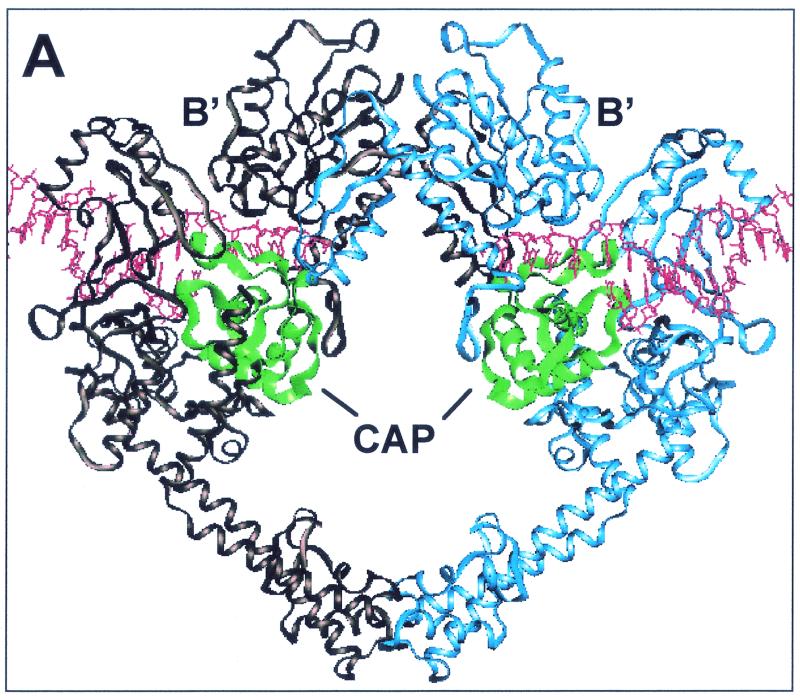
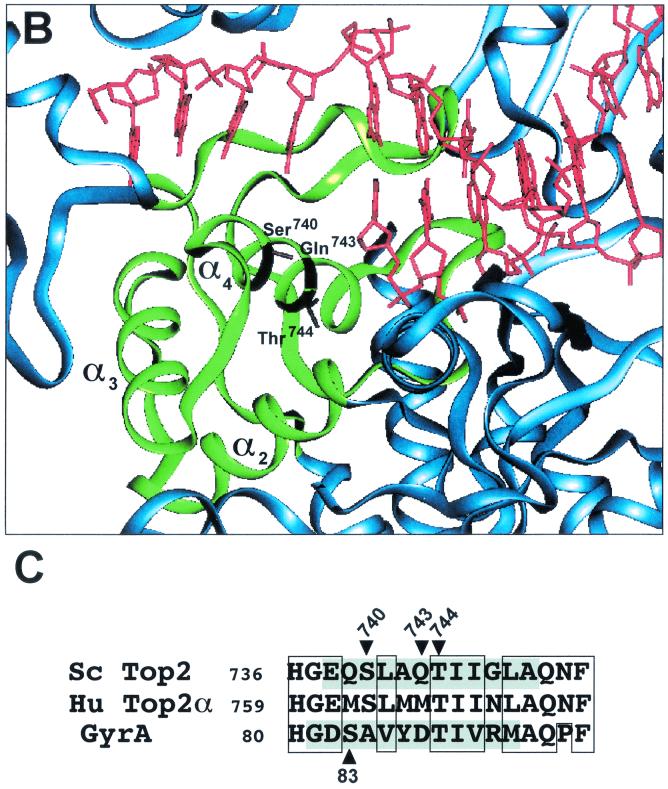
FIG. 1.
CAP homology domain and positions of Ser740, Gln743, and Thr744 in yeast Top2p. (A) Illustration of dimeric S. cerevisiae topoisomerase II structure (Protein Data Bank accession number 1bgw). Drawings were generated using the program QUANTA (version 97). The helical ribbon representation shows the 92-kDa fragment of the yeast enzyme with a DNA fragment modeled into each of the putative DNA-binding sites (6). The CAP homology domain of each protomer is highlighted in green. (B) Close view of the putative DNA-binding region, presenting the proposed proximity of the α4-helix within the CAP homology domain, including Ser 740, Gln 743, and Thr744 (in a stick model), to DNA. (C) Alignment of protein sequence for the yeast (Sc Top2p), human Top2pα (Hu Top2pα) and E. coli gyrase. The mutated residues studied in the present report are indicated by arrowheads. Shaded residues correspond to the α4-helix (5). No shading is shown for human Top2p because of lack of structural data. Boxed regions indicate similarity between the amino acid residues. Ser763 in the human Top2pα sequence corresponds to Ser740 in yeast Top2p.