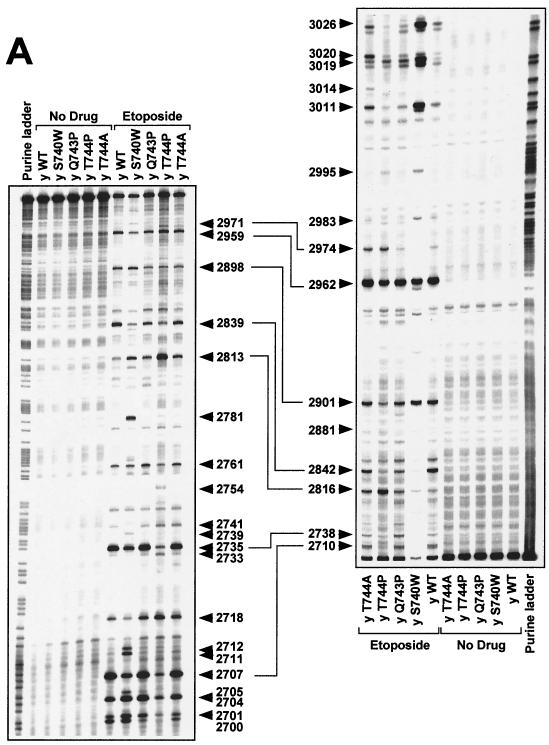
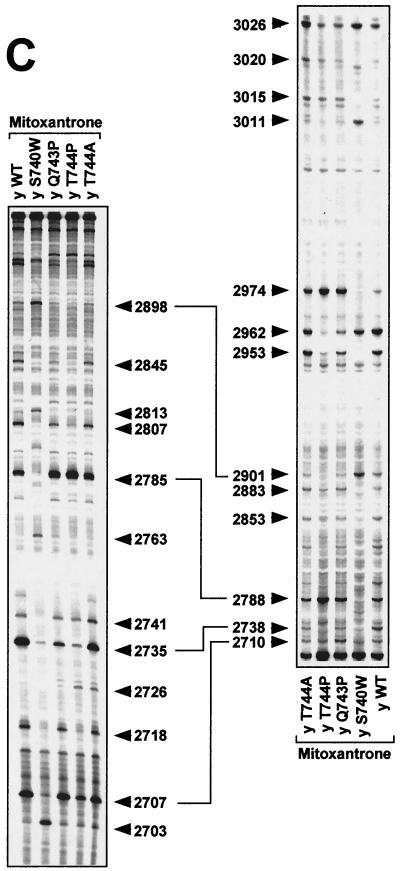
FIG. 3.
DNA cleavage patterns generated by yeast Top2pWT, Top2pS740W, Top2pQ743P, Top2pT744P, and Top2pT744A enzymes in the presence of etoposide, the fluoroquinolone CP-115,953, and the intercalator mitoxantrone. DNA fragments from the junction between the c-myc first intron and first exon between positions 2671 and 3072 were prepared by PCR using one primer labeled with 32P at the 5′ terminus. The left-hand panels show labeling of the upper DNA strand at position 2671. The right-hand panels show that labeling of the lower DNA strand was at position 3072. Top2 enzymes are indicated above each lane. Lanes: y WT, yeast wild-type Top2p; y S740W, yeast Top2pS740W; y Q743P, yeast Top2pQ743P; y T744P, yeast Top2pT744P; y T744A, yeast Top2pT744A. Concentrations were 100 μM for etoposide (A) and CP-115,953 (B) and 1 μM for mitoxantrone (C). Reactions were performed at 37°C for 30 min in the presence of 5 mM MgCl2. Purine ladders were obtained after formic acid reaction. Numbers correspond to genomic positions of the nucleotide covalently linked to Top2p via the 5′ phosphate. Double-headed arrows correspond to DNA cleavage sites with a 4-bp stagger that represent potential DNA double-strand breaks.