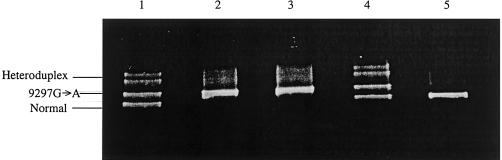
Figure 2.
Identification of AGAT nonsense mutation in lymphoblasts of two female siblings and leukocytes of their parents, by DGGE analysis of exon 3. Lane 1, Father, genotype T149X/N. Lane 2, Patient 1, genotype T149X/T149X. Lane 3, Patient 2, genotype T149X/T149X. Lane 4, Mother, genotype T149X/N. Lane 5, Wild-type DNA from unaffected control, genotype N/N. DNA extraction from EBV-transfected lymphoblasts and cultivated fibroblasts was performed using guanidinhydrochloride/proteinase K (Nucleospin C+T kit [Machery Nagel]). PCR primers were designed to cover exons 1–9 and the adjoining introns of the AGAT gene. PCR reactions were carried out over 37 cycles with 2.5 U of Ampli-Taq Gold DNA polymerase (Perkin Elmer) in a Perkin-Elmer-Cetus DNA Thermal Cycler. DGGE analysis of PCR products was essentially performed as described by Greber-Platzer et al. (1997), after heteroduplex formation by loading 20 μl of sample onto an 8% polyacrylamide gel containing a 15%–55% or 25%–65% linearly increasing denaturing gradient (100% = 7 M urea and 40% (vol/vol) deionized formamide). Electrophoresis was performed at 160 V for 6 h in a 1× TAE buffer at 60°C, using a DCode system (Biorad). After electrophoresis, gels were stained in ethidium bromide and examined under ultraviolet illumination. DGGE analysis of exon 1 was not possible, because of its high melting temperature; therefore, direct sequencing of exon 1 was performed. In the patients, no aberrations from the normal sequence were found on exon 1.