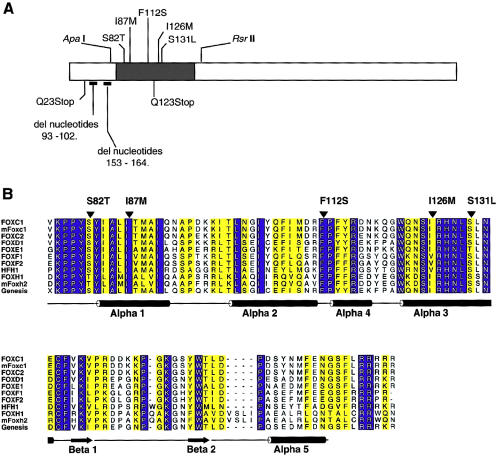
Figure 2.
A, Schematic of the FOXC1 protein. The blackened rectangle represents the forkhead domain, and the missense mutations studied are indicated above the forkhead domain. Mutations presented below the ideogram result in truncated products and were not tested. Positions of restriction-enzyme sites used in subcloning are indicated above the ideogram. B, Multiple-sequence alignment of the forkhead domain of human FOXC1 and related FOX proteins. The sequences shown in single-letter amino acid codes are those of human FOXC1 (AF048693), mouse Foxc1 (NM008592), human FOXC2 (Y08223), human FOXD1 (U13222), human FOXE1 (U89995), human FOXF1 (U13219), human FOXF2 (U13220), human HFH1 (AF153341), human FOXH1 (AF076292), mouse Foxh2 (AF110506), and rat Genesis/Foxd3 (NM012183), respectively (NCBI Databases) . Amino acid residues showing absolute identity among these proteins are shown in white against a blue background; those positions with conservative substitutions are shown against a yellow background. The positions of the α-helices and β-strands, as defined in the NMR structure of Genesis, are schematically represented below the alignment. Positions of mutations and corresponding amino acid changes in FOXC1 are indicated by the blackened arrowheads above the alignment. ALSCRIPT was used to format the alignment.