Abstract
A key complication in comparative genomics for reliable gene function prediction is the existence of duplicated genes. To study the effect of gene duplication on function prediction, we analyze orthologs between pairs of genomes where in one genome the orthologous gene has duplicated after the speciation of the two genomes (i.e. inparalogs). For these duplicated genes we investigate whether the gene that is most similar on the sequence level is also the gene that has retained the ancestral gene-neighborhood. Although the majority of investigated cases show a consistent pattern between sequence similarity and gene-neighborhood conservation, a substantial fraction, 29–38%, is inconsistent. The observation of inconsistency is not the result of a chance outcome owing to a lack of divergence time between inparalogs, but rather it seems to be the result of a chance outcome caused by very similar rates of sequence evolution of both inparalogs relative to their ortholog. If one-to-one orthologous relationships are required, it is advisable to combine contextual information (i.e. gene-neighborhood in prokaryotes and co-expression in eukaryotes) with protein sequence information to predict the most probable functional equivalent ortholog in the presence of inparalogs.
INTRODUCTION
Comparative genomics has become an important research area in the wake of the large number of sequenced genomes that have become available in recent years. The comparison of genomes is of great importance in the prediction of gene function and to study the evolution of genomic properties such as gene-neighborhood. These comparative studies rely on homology and increasingly on orthology, because orthologs originated from a single gene in the last common ancestor by speciation (1).
The bidirectional best hit (BBH) method is a widely used homology based procedure for orthology that, in general, results in a single gene in one genome being predicted to be the ortholog of a single gene in the other genome. The BBH method has been applied in various function prediction studies, such as the construction of a conserved co-expression network and the prediction of regulatory motifs (2,3). However, one major complication in the BBH method exists when gene duplication events have occurred after the speciation of the two genomes under investigation. To distinguish these gene duplicates from more ancient and hence presumably more functionally diverged duplicates, Sonnhammer and Koonin (4) coined the phrase inparalogs for duplicated genes after a speciation event. Both these inparalogs are then orthologous to a single gene in the other species (referred as co-orthology). By only using BBH many of such truly orthologous relations will not be detected, and hence computational methods have been developed to include inparalogs (5,6). Despite the availability of methods that include inparalogs, scientists often use these programs in such a way that genes (still) only have one ortholog, thereby effectively resorting to BBH-like heuristics and ignoring small differences in sequence similarity of inparalogs to their ortholog (7). The intuitive idea behind this approach seems logical, because duplication can lead to a differentiation process in which only one of the two inparalogs retains the ancestral function. It is expected that in such cases the most similar inparalog (to the single-ortholog) is the one that has retained that ancestral function. However, especially for small differences in sequence similarity in a large sequence space, this intuitive idea may not be correct: both inparalogs, or even only the less similar one, might carry out the ancestral function.
In order to study whether or not inparalogs have retained the (ancestral) function, we can use methods that map gene function on a genome-wide scale, such as co-expression or genomic context methods (8–10). Although these methods are indicators of biological process (11), rather than molecular function, it gives useful insights into the function of recently duplicated genes. For example, in a study that measured the conservation of co-expression, inparalogous genes were detected to often have diverged in terms of their co-expression: one of the duplicates retained co-expression, while the other did not (12). Here, we use gene-neighborhood conservation as the genomic context method, to study the relationship between gene function and sequence evolution of recent gene duplicates. Gene-neighborhood provides very strong signals for functional association between gene products within and between species (8,13–15).
In this analysis we addressed whether genes that are the most similar on the sequence level are also the ones that have retained the ancestral gene-neighborhood and hence are likely to function in the same biological process as their ortholog. Surprisingly, we have found in 29–38% of investigated co-ortholog relationships that the less similar gene pair retained the ancestral gene-neighborhood. Therefore, the BBH does not necessarily correspond to contextual information (biological process). Although, the majority of cases show a consistency between BBH and gene-neighborhood conservation, it is advisable to combine contextual information with protein sequence information to predict the most probable functional equivalent ortholog in the presence of inparalogs.
METHODS
Co-orthology detection
Our approach to investigate if there is a relationship between protein sequence similarity and gene-neighborhood conservation is based on co-orthology detection by the Inparanoid algorithm, using the default settings. Inparanoid constructs co-ortholog groups by applying a specific clustering algorithm to assign inparalogs to existing BBH pairs. This clustering algorithm is based on the assumption that inparalogs are more similar to each other than to any other sequence from the other genome (5).
Data set of genomes
We used the Genbank genomes of species at varying phylogenetic distances, including four Archaea (Archaeoglobus fulgidus DSM 4304, Halobacterium sp. NRC-1, Methanocaldococcus jannaschii DSM 2661 and Sulfolobus solfataricus P2), four Gram-positive bacteria (Lactobacillus plantarum WCFS1, Lactococcus lactis IL1403, Bacillus cereus ATCC 14579 and Bacillus subtilis subsp. subtilis str. 168) and four Proteobacteria (Escherichia coli K12, Pseudomonas aeruginos PAO1, Helicobacter pylori J99 and Caulobacter crescentus CB15). The genome information was taken from the NCBI database (ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/).
Gene-neighborhood conservation for function prediction
We used gene-neighborhood conservation to predict functional equivalency (with regard to biological process) between orthologs. For each ortholog pair we parsed the Genbank files to obtain two clusters of genes (one for each ortholog), consisting of three genes 5′ and three genes 3′ of the query ortholog. We then counted the total number of ortholog pairs present in both clusters, resulting in a score scaled from 0 (no conservation) to 6 (complete conservation), irrespective of the precise gene order or the relative direction of transcription.
Dataset of co-ortholog groups and comparison of gene-neighborhood conservation
In order to measure the evolution of gene-neighborhood relative to the sequence evolution we specifically extracted the two-to-one (2:1) co-ortholog groups from the Inparanoid outputs (for the total number of many-to-many orthologous relationships see Table 1). These groups consist of a BBH and one additional inparalog, which is the second best hit (SBH) of the unique gene from the other genome (single-ortholog) (Figure 1). Subsequently, the number of neighboring orthologs for each ortholog pair within the co-ortholog group was compared. This comparison was done for all 2:1 co-ortholog groups obtained from several pairwise genome-comparison datasets; (i) Gram-positive bacteria, (ii) Proteobacteria, (iii) Archaea, (iv) Gram-positive bacteria and Proteobacteria, (v) Gram-positive bacteria and Archaea, (vi) Proteobacteria and Archaea and (vii) Gram-positive bacteria, Proteobacteria and Archaea.
Table 1.
Number of x-to-y pairwise orthologous relationships in total dataset
| x/y | 1 | 2 | 3 | 4 | 5 | ≥6 |
|---|---|---|---|---|---|---|
| 1 | 38 560 | 5072 | 923 | 291 | 110 | 124 |
| 2 | 287 | 176 | 55 | 28 | 41 | |
| 3 | 18 | 21 | 9 | 24 | ||
| 4 | 3 | 7 | 11 | |||
| 5 | 0 | 11 | ||||
| ≥6 | 8 |
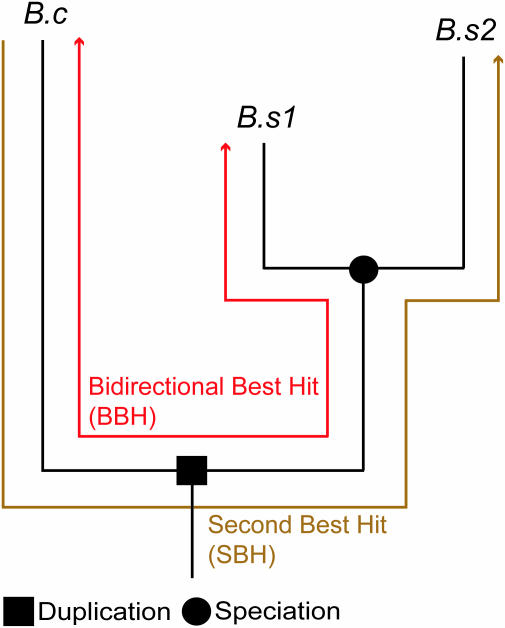
Figure 1.
Two-to-one (2:1) co-ortholog relationship between Bacillus cereus (B.c) and Bacillus subtilis (B.s). The number of neighboring orthologs for the BBH and the SBH were compared.
RESULTS
Consistency and inconsistency between sequence similarity and gene-neighborhood conservation
To measure the evolution of inparalogs, within two-to-one co-ortholog groups, we classified and counted the occurrences of various possible evolutionary outcomes in terms of gene-neighborhood conservation and sequence similarity (Figure 2). We are specifically interested in those cases where one of the two inparalogs has lost all traces of the ancestral gene-neighborhood while the other still retains it. We expect that the inparalog that has retained the ancestral gene-neighborhood is the preferred copy for the biological process that this chromosomal gene cluster performs. Such cases raise the question how the relative sequence similarity of the inparalogs (which is an important parameter in orthology detection) relates to retaining the ancestral gene-neighborhood. In order to analyze this, we refer to the inparalog with the highest similarity as the BBH and the inparalog with the lowest similarity as the SBH (of the single-ortholog). Inconsistencies are then defined as cases where the SBH has a conserved gene-neighborhood while at the same time the BBH does not and vice versa for the consistencies. Although the majority of investigated cases show a consistent pattern between sequence similarity and gene-neighborhood conservation, a substantial fraction, 29–38%, does not (Figure 3).
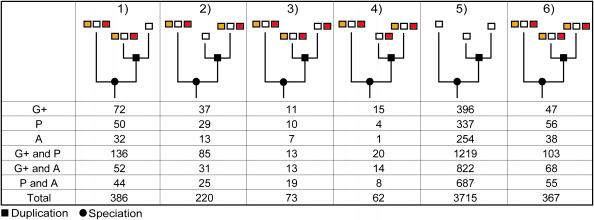
Figure 2.
Number of co-ortholog groups per class for several genome-comparison data sets; 1, only the BBH has a conserved gene-neighborhood; 2, only the SBH has a conserved gene-neighborhood; 3, unequal number of conserved neighboring genes, but BBH conserves a higher number; 4, unequal number of conserved neighboring genes, but SBH conserves a higher number; 5, no gene-neighborhood conservation; and 6, equal number of conserved neighboring genes. The genome-comparison datasets include species from Gram-positive bacteria (G+), Proteobacteria (P) and Archaea (A). Note that the 2:1 co-ortholog relationships in which the inparalogs are 100% identical on the sequence level are excluded (owing to BBH and SBH definition).
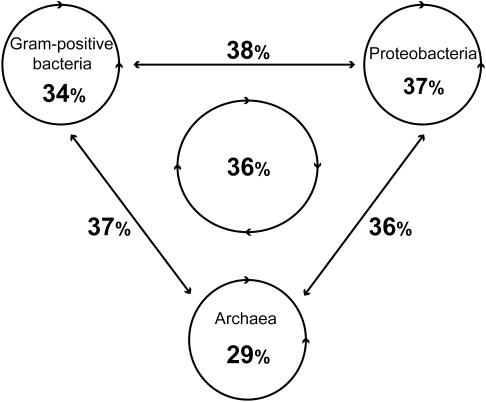
Figure 3.
Percentage of inconsistency for the several genome-comparison datasets. An inconsistency is found when the SBH (sequences with relatively the lowest sequence similarity) has a conserved gene-neighborhood in contrast to the BBH.
Although the number of observed inconsistencies varies between the several genome-comparison datasets, the percentages of inconsistencies are similar. Therefore, inconsistencies are not limited to specific genome comparisons. Furthermore, consistencies and inconsistencies are also found in which both BBH and SBH have a conserved gene-neighborhood, but with unequal numbers (Figure 2, class 3 and 4). Because the total number of such cases is lower, the variance in the percentage of inconsistencies is larger: 13–61%.
Inconsistencies are not caused by inparalog detection artifacts
It is known that relative BLAST hits do not necessarily reflect the actual evolutionary history of genes. One type of event that is likely to be a problem is an ancient gene duplication which has taken place before speciation, resulting in what is referred to as outparalogs (4). It is possible that BLAST hit driven methods call two genes co-orthologous to a single-ortholog (thus inparalogs), while in fact, according to phylogenetic tree reconstruction, one of them is an outparalog and the other is a real one-to-one ortholog (Figure 4). This kind of difference between relative BLAST hits and trees are to a certain level expected because both methods are based on different approaches. For example, Inparanoid (BLAST hit driven approach) has been specifically designed for large-scale pairwise genome comparisons, based on relatively little sequence information. In contrast, more sequence information from multiple species is used in phylogenetic tree reconstruction, but reliable (large-scale) automatic procedures are not yet available. We performed several phylogenetic tree reconstructions to check if our cases of inconsistency could possibly be due to ‘relative BLAST hit’ artifacts. COG, MUSCLE and PHYML were used for the phylogenetic tree reconstructions (for details see Figure 4) (6,16,17). We tested an equal number of randomly selected consistencies and inconsistencies from the total dataset to confirm whether the recent gene duplicates are also inparalogs according to the phylogenetic tree. The number of confirmed inparalogs from the test set appeared to be high (85%) and, more importantly, almost equally distributed among consistent and inconsistent cases (9 and 8 out of 10, respectively). The inconsistencies are thus not likely to be the result of deficiencies of our applied large-scale method (Inparanoid).
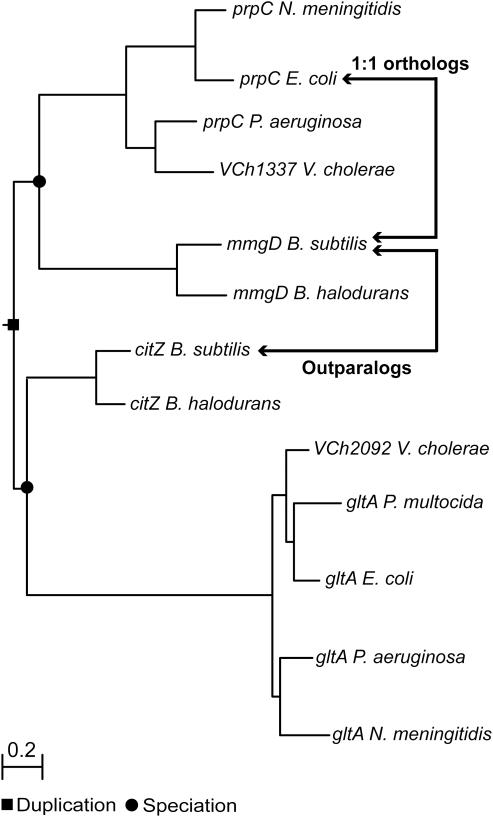
Figure 4.
Example of an outparalog relationship. Two paralogs from B.subtilis (mmgD and citZ) are inparalogs relative to prpC of E.coli, according to Inparanoid. However, the phylogenetic tree shows an outparalog relationship between mmgD and citZ, because the duplication event took place before speciation to B.subtilis and E.coli. Therefore, citZ is not orthologous to prpC. To construct the phylogenetic tree, we first selected from the COG database the specific COG to which mmgD, citZ and prpC belong (6). All homologs, included in this specific COG, were used as input for MUSCLE to construct a multiple sequence alignment (16). Finally, the phylogenetic tree was constructed, from the multiple sequence alignment, using the PHYML algorithm (17).
Inconsistencies are not caused by a lack of divergence time
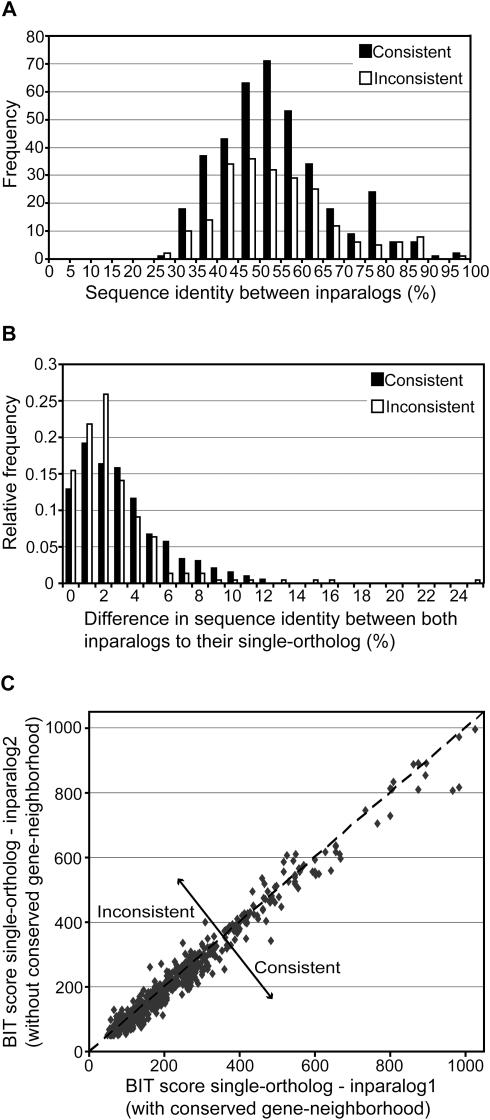
To analyze the possible cause of the inconsistencies, we have investigated the percentage of sequence identity between in paralogs within consistencies and inconsistencies. This sequence identity is a measure of how recent the duplication events are. If the inparalogs are highly identical on the amino acid sequence level, the inconsistencies could be observed because of a chance difference owing to a lack of sufficient divergence time. The analysis reveals that our observation of inconsistencies is not the result of this particular explanation, because the majority of the inparalogs are diverged to a high extent (Figure 5A). In fact, for the few cases of recently duplicated genes the ratio between consistency and inconsistency is closer to 50%, supporting the prediction that a lack of sufficient divergence time between the duplicates can cause inconsistencies by chance. The observed high level of divergence between duplicated genes could suggest a possible functional differentiation with respect to their biological process and/or molecular function (18).
Figure 5.
Sequence identity analysis between inparalogs. All plots are constructed from the total genome-comparison dataset, including Gram-positive bacteria, Proteobacteria and Archaea. (A) Frequency of consistent (most similar inparalog has a conserved gene-neighborhood) and inconsistent cases (less similar inparalog has a conserved gene-neighborhood) at different levels of sequence identity between inparalogs. Nine co-ortholog groups were found in which the inparalogs show 100% sequence identity. These are not included in this plot, because both inparalogs are in fact BBHs. Therefore, a consistency or inconsistency, as defined, cannot be determined. (B) Relative frequency of consistent and inconsistent cases at different levels of sequence identity difference between inparalogs to their single-ortholog. The sequence identity difference is calculated by subtracting the percentage of sequence identity between the BBH and the SBH. (C) Sequence similarity, expressed in BIT scores, between the single-ortholog and both inparalogs. The inparalog which is located in a conserved gene-neighborhood is plotted on the X-axis. Therefore, the consistencies are positioned underneath the line y = x.
Sequence evolution of inparalogs relative to their single-ortholog
An interesting question regarding the inparalogs is whether or not gene duplicates are differentiated with respect to their molecular function, but more importantly, if this is different for consistencies and inconsistencies. Although no direct measurement of molecular function is available on a genomic scale, we can indirectly investigate this from the sequence divergence of inparalogs relative to their single-ortholog in the other genome. Figure 5B and C shows that within the consistent and inconsistent cases both inparalogs are of comparable similarity to their single-ortholog. We thus observe little asymmetry in the rate of sequence evolution between gene duplicates, which has also been described in previous studies (18,19). As Inparanoid is not, in general, able to detect inparalogs with large sequence differences (to their single-ortholog), a small difference is to a certain level to be expected. However, we observed a tendency towards an even smaller difference between inconsistent inparalogs (Figure 5B), suggesting that the inconsistent inparalogs have retained the same molecular function. This is supported by the fact that the majority of accepted amino acid substitutions in sequence evolution are only subject to purifying (or negative) selection while very few substitutions are positively selected and alter the molecular function of a gene (such as co-factor preference) (20). In spite of this, it cannot completely be excluded that a change in molecular function has occurred. Such a change normally only depends on a small number of amino acid substitutions (20), which easily remains undetected against the background of many substitutions (at least with methods like BLAST). In the following sections we will show three examples of functional differentiation between inconsistent inparalogs that have been partly experimentally characterized.
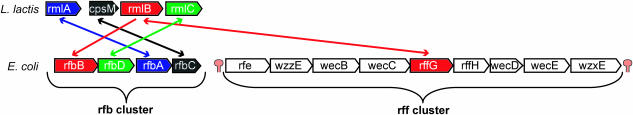
Rfb and rff gene cluster
The E.coli genome contains the two gene clusters (operons) rfb and rff, each containing one member of the inparalogous pair rfbB and rffG (Figure 6). RfbB and RffG are known to both catalyze the same biochemical reaction (dTDP-d-glucose ↔ H2O + dTDP-4-dehydro-6-deoxy-d-glucose), but are involved in two different biological processes (21). Genes in the rfb gene cluster (genes: rfbB, rfbD, rfbA and rfbC) are involved in the biosynthesis of O-specific polysaccharides, which are components of the membrane-localized lipopolysaccharide. In contrast, genes in the rff gene cluster are involved in the complex biosynthesis of enterobacteria common antigen, which is located in the outer membrane. L.lactis contains only the rfb gene cluster (genes: rmlA, cpsM, rmlB and rmlC). E.coli rffG (part of the rff gene cluster) and L.lactis rmlB (part of the rfb gene cluster) are BBHs, despite the fact that they are part of two different biological processes. In fact, E.coli rfbB and L.lactis rmlB (SBH pair) are the most reliable functional equivalents, because these two genes have a conserved gene-neighborhood (Figure 6, rfb gene cluster). In this example, the BBH does not represent the functional equivalent in terms of biological process.
Figure 6.
Genome comparison between E.coli and L.lactis: the rfb and rff gene cluster. The rff gene cluster is not found in L.lactis by the applied ortholog detection method (Inparanoid). Bidirectional and unidirectional arrows indicate a BBH and a SBH, respectively. The rff gene cluster is directly flanked by transcription termination signals [predicted by Transterm (34)].
Gab gene cluster
Both E.coli and B.cereus contain the gene gabT, which codes for a 4-aminobutyrate transaminase. This enzyme is part of the 4-aminobutyrate-degradation pathway. A neighboring gene of gabT on the E.coli and B.cereus genome is gabD, which is also necessary to degrade 4-aminobutyrate to succinate. Both genes are under a complex regulation process, induced by stress conditions, and are reported to be co-transcribed (22,23). However, gabT of B.cereus and goaG of E.coli are BBHs in a pairwise genome comparison. GoaG is not known to be involved in a biochemical reaction or pathway and it does not have conserved neighboring genes on the genome. In contrast, the gabT genes of E.coli and B.cereus are detected as a SBH pair, but they are both flanked by gabD. It is clear that the SBH, instead of the BBH, represents the most probable functional equivalents.
Prp gene cluster
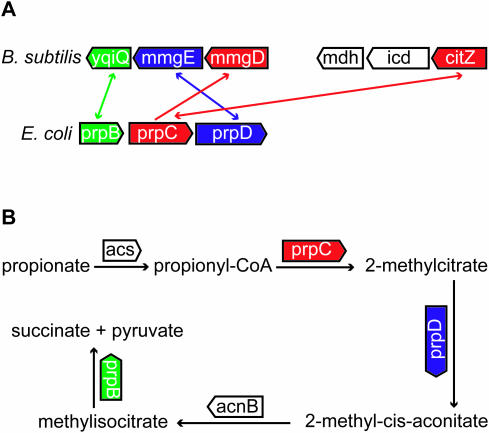
The prp gene cluster in E.coli codes for enzymes involved in the methylcitrate cycle, in which propionate is degraded to pyruvate and succinate (Figure 7). The cluster contains prpC that codes for a 2-methylcitrate synthase. From biochemical studies it is known that PrpC has affinity for both acetyl-CoA and propionyl-CoA. Propionyl-CoA is converted to 2-methylcitrate by 2-methylcitrate synthase (EC 4.1.3.31), whereas acetyl-CoA is converted to citrate in the citric acid cycle by citrate synthase (EC 2.3.3.1). Moreover, it has been shown that 2-methylcitrate synthase and citrate synthase are regulated independently, because 2-methylcitrate synthase is only activated during growth on propionate, thereby supporting a functional difference (24).
Figure 7.
(A) Co-orthology relationship between prpC of E.coli, mmgD and citZ of B.subtilis. mmgD is the SBH of prpC (unidirectional arrow), whereas citZ and prpC are BBHs (bidirectional arrow). prpC and mmgD show, in contrast to the BBH, conservation of neighboring genes, which are all involved in the methylcitrate cycle. citZ is part of the citric acid cycle and is therefore not functional equivalent to prpC, for details see text. (B) The neighboring genes prpB, prpC and prpD are indicated on the biochemical map of the methylcitrate cycle. The metabolic information is taken from Ecocyc (http://ecocyc.org/).
However, two inparalogs of B.subtilis (citZ as part of a BBH and mmgD as SBH) are found in a comparison with E.coli. The citZ gene is annotated and experimentally verified as a citrate synthase (25). MmgD is also annotated as a citrate synthase, but there is no experimental evidence. The BBH does not show any gene-neighborhood conservation, whereas the SBH does. A functional equivalency between prpC and mmgD, instead of prpC and citZ, is therefore very probable (Figure 7A and B). Moreover, icd and mdh are direct neighbors of citZ and are annotated as isocitrate dehydrogenase and malate dehydrogenase, respectively. The metabolic product of malate dehydrogenase is oxaloacetate, which is the substrate of CitZ. Furthermore, the metabolic product of isocitrate dehydrogenase, α-ketoglutarate, is known to be a competitive inhibitor of CitZ (26). This biochemical knowledge clearly indicates a functional association between the three neighboring genes, citZ, icd and mdh, as part of the citric acid cycle.
Sequence alignment between the inparalogs, citZ and mmgD, shows a 43% sequence identity. This is an indication that the duplication event was not very recent, so that there was actually time for functional differentiation of the inparalogs (CitZ as part of the citric acid cycle instead of the methylcitrate cycle). Careful examination of a phylogenetic tree of this homologous family revealed that there is an outparalog relationship between citZ and mmgD (event of gene duplication took place before speciation to E.coli and B.subtilis) rather than an inparalog relationship proposed by the Inparanoid method (Figure 4). This example shows not only an inconsistent pattern between sequence similarity and gene-neighborhood conservation, but also inconsistency between relative BLAST hits and phylogenetic tree reconstruction. In fact, the tree indicates a one-to-one ortholog relationship between mmgD and prpC, which are according to gene-neighborhood conservation also the most probable functional equivalents. This supports our message that protein sequence information should be combined with contextual information to predict the true functional equivalents in cases of inparalogs, especially when these are obtained from ‘relative BLAST hit’ methods.
DISCUSSION
One-to-one orthology versus gene-neighborhood conservation
We have studied how often the functional equivalents, according to gene-neighborhood conservation, are also the genes that are the most similar at the amino acid sequence level. Although in many cases there is conservation of gene-neighborhood between the detected BBHs (in the presence of inparalogs), it is surprisingly not true in all cases. We have found in 29–38% of the cases that only the less similar ortholog pair (SBH) has a conserved gene-neighborhood. Therefore, in a substantial fraction, the most probable functional equivalents are the genes in an SBH pair instead of the BBH.
Evolutionary tracks of gene duplicates in terms of biological process
As most of the duplication events are not recent (Figure 5A), the duplicates are expected to have differentiated to some degree as is also suggested by the differential retention of the ancestral gene-neighborhood. Specifically, it is interesting whether one of the gene duplicates is now active in a completely distinct biological process (i.e. neofunctionalization with respect to the process) or whether it is still active in the same biological process [i.e. subfunctionalization within the process (27,28)]. We therefore investigated the new genomic context of ‘inconsistent’ inparalogs in E.coli in relation to the ancestral context as defined by the single-ortholog. We took E.coli because the transcriptional organization of its genome is well characterized (e.g. operons). Seventeen inconsistent cases were amenable to analysis. In 10 cases, both ‘inconsistent’ inparalogs are part of two different operons which are active in the same general biological process, according to the COG functional classification. In contrast, only two ‘inconsistent’ inparalogs are in a context that clearly indicates a different biological process: the members of one operon are classified in different COG functional categories compared with the members of the second operon. Most inconsistent inparalogs seem thus to have undergone subfunctionalization rather than neofunctionalization. The remaining five cases included inparalogs which were classified as a transcriptional unit consisting of one gene. We suspect that these isolated inparalogs are still functionally associated to their ancestral components, but active under specific cellular conditions. The importance of such differential regulation (activation under different conditions) of genes that catalyze similar reactions in large-scale biological networks have been shown previously by Ihmels et al. (29). We have, for example, observed individual genes, such as substrate-binding proteins of ATP-binding cassette (ABC) transporter systems, which do not reside in a gene cluster with other essential ABC transporter components. It is possible that such a solitary substrate-binding inparalog codes for a protein with slightly altered substrate specificity, but which still uses the ancestral ABC transport system to transport external substrates (e.g. Supplementary Data) under specific conditions.
Why do we observe inconsistencies?
It is very likely, and for a number of cases we have documented, that inparalogs which have lost the ancestral gene-neighborhood are not the preferred copy for their ancestral process and have subfunctionalized within the same general process or have even neofunctionalized. However, the gene duplicates with a different context can be more similar to the single-ortholog on the sequence level, which raises the questions: what does this imply for the molecular function of ‘inconsistent’ inparalogs and why do we observe these inconsistencies? As we observe little difference of inparalogs to their single-ortholog and an even smaller difference for the inconsistent inparalogs, it suggests that both inparalogs have a very similar molecular function. Nevertheless, it cannot be excluded that substantial changes in molecular function have occurred in the evolution of these inconsistent inparalogs, given that only one amino acid substitution can change, e.g. the substrate specificity of an enzyme (20). A change in the molecular function of inconsistent inparalogs could negatively contribute to the fitness of a given species (because the inparalog is still in the conserved gene-neighborhood), which would imply that more inconsistencies are observed for populations with a low or even a negative selection co-efficient. We tested this effect by comparing the inconsistency percentage to the ‘selected codon usage bias’ as a measure of the strength of selection in that species (30). To our surprise we did not find this effect. Instead there is no observable correlation between selection strength and the percentage of inconsistency, although we might have too few species to detect any such trend even if it exists (Supplementary Data). Given our inability to detect any correlation, let alone a significant negative correlation, it seems likely that most of our inparalogs have a very similar molecular function. Any inconsistencies are then a chance outcome: both duplicates have diverged, but at (roughly) the same evolutionary speed (Figure 5B and C). Such a similar rate of sequence evolution has been demonstrated previously, at least for recent gene duplicates (18), and occurs because most amino acids substitutions have only been subject to purifying selection and not to adaptive selection (20).
Implication of co-orthology on function prediction
Our large-scale analysis and the experimentally characterized cases confirm that orthology detection by BBH can negatively influence function predictions when gene-neighborhood conservation and co-orthology are not considered. The occurrence of inparalogs is an issue for at least 16% of the genes in pairwise species comparisons (Table 1), but it is even more important for group orthology schemes, such as COG, where virtually no orthologous group consists of only one gene per species. The importance of including inparalogs was also recently shown for increasing the accuracy in operon predictions (31). However, the evolutionary fate of the investigated inparalogs with respect to their ancestral function is still not completely known [several possible functional diversification scenarios do exist, as recently discussed by Hughes (27)]. Therefore, in line with the original (evolutionary) definition of orthology one should include inparalogs and take both genes as equally probable candidates for function prediction. If one insists to pinpoint a functional equivalent inparalog, gene-neighborhood should be combined with protein sequence conservation. By such a combination one should take into account that the inparalog which is located in a conserved gene-neighborhood is likely to be the preferred copy for the ancestral process. In cases where gene-neighborhood conservation is absent (like in eukaryotes) the general principle of contextual information can still be applied to increase the accuracy of function prediction. There is sufficient contextual information available for eukaryotes, e.g. co-expression datasets and predicted transcription factor-binding sites. Other types of functional information are interactions from proteomics, evolutionary conservation of gene fusion and literature mining. In fact, some of these other types of contextual information (i.e. protein–protein interactions and microarray derived co-expression) have been used recently to study the functional differentiation of duplicated genes in eukaryotes (32,33). As a result of many upcoming functional genomic studies, including genome sequencing projects as well as high-throughput co-expression and protein–protein interaction analysis, the importance of contextual information in gene function prediction will rapidly increase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Jos Boekhorst, Robert Kerkhoven, Bas Dutilh and Christof Francke for stimulating discussions and we thank Paul Sharp for providing unpublished data on the strength of selected codon usage bias for archaea. This work was part of (i) The BioRange programme of The Netherlands Bioinformatics Centre (NBIC), which is supported by a BSIK grant through The Netherlands Genomics Initiative (NGI) and (ii) The IOP Genomics grant IGE01018. Funding to pay the Open Access publication charges for this article was provided by the IOP Genomics grant nr fn6877.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fitch W.M. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 2.Stuart J.M., Segal E., Koller D., Kim S.K. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 3.Pritsker M., Liu Y.C., Beer M.A., Tavazoie S. Whole-genome discovery of transcription factor binding sites by network-level conservation. Genome Res. 2004;14:99–108. doi: 10.1101/gr.1739204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonnhammer E.L.L., Koonin E.V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 5.Remm M., Storm C.E.V., Sonnhammer E.L.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 6.Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 7.Wuchty S., Almaas E. Peeling the yeast protein network. Proteomics. 2005;5:444–449. doi: 10.1002/pmic.200400962. [DOI] [PubMed] [Google Scholar]
- 8.Dandekar T., Snel B., Huynen M., Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 9.Marcotte E.M., Pellegrini M., Ng H.L., Rice D.W., Yeates T.O., Eisenberg D. Detecting protein function and protein–protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 10.Enright A.J., Iliopoulos I., Kyrpides N.C., Ouzounis C.A. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 11.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snel B., van Noort V., Huynen M.A. Gene co-regulation is highly conserved in the evolution of eukaryotes and prokaryotes. Nucleic Acids Res. 2004;32:4725–4731. doi: 10.1093/nar/gkh815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overbeek R., Fonstein M., D'Souza M., Pusch G.D., Maltsev N. Use of contiguity on the chromosome to predict functional coupling. In Silico Biol. 1998;1:93–108. [PubMed] [Google Scholar]
- 14.Overbeek R., Fonstein M., D'Souza M., Pusch G.D., Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snel B., Lehmann G., Bork P., Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 18.Kondrashov F.A., Rogozin I.B., Wolf Y.I., Koonin E.V. Selection in the evolution of gene duplications. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-research0008. RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kellis M., Birren B.W., Lander E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 20.Golding G.B., Dean A.M. The structural basis of molecular adaptation. Mol. Biol. Evol. 1998;15:355–369. doi: 10.1093/oxfordjournals.molbev.a025932. [DOI] [PubMed] [Google Scholar]
- 21.Marolda C.L., Valvano M.A. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J. Bacteriol. 1995;177:5539–5546. doi: 10.1128/jb.177.19.5539-5546.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartsch K., von Johnn-Marteville A., Schulz A. Molecular analysis of two genes of the Escherichia coli gab cluster: nucleotide sequence of the glutamate:succinic semialdehyde transaminase gene (gabT) and characterization of the succinic semialdehyde dehydrogenase gene (gabD) J. Bacteriol. 1990;172:7035–7042. doi: 10.1128/jb.172.12.7035-7042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzner M., Germer J., Hengge R. Multiple stress signal integration in the regulation of the complex sigma S-dependent csiD-ygaF-gabDTP operon in Escherichia coli. Mol. Microbiol. 2004;51:799–811. doi: 10.1046/j.1365-2958.2003.03867.x. [DOI] [PubMed] [Google Scholar]
- 24.Gerike U., Hough D.W., Russell N.J., Dyall-Smith M.L., Danson M.J. Citrate synthase and 2-methylcitrate synthase: structural, functional and evolutionary relationships. Microbiology. 1998;144:929–935. doi: 10.1099/00221287-144-4-929. [DOI] [PubMed] [Google Scholar]
- 25.Jin S., Sonenshein A.L. Identification of two distinct Bacillus subtilis citrate synthase genes. J. Bacteriol. 1994;176:4669–4679. doi: 10.1128/jb.176.15.4669-4679.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira D.S., Donald L.J., Hosfield D.J., Duckworth H.W. Active site mutants of Escherichia coli citrate synthase. Effects of mutations on catalytic and allosteric properties. J. Biol. Chem. 1994;269:412–417. [PubMed] [Google Scholar]
- 27.Hughes A.L. Gene duplication and the origin of novel proteins. Proc. Natl Acad. Sci. USA. 2005;102:8791–8792. doi: 10.1073/pnas.0503922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch M., Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ihmels J., Levy R., Barkai N. Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat. Biotechnol. 2004;22:86–92. doi: 10.1038/nbt918. [DOI] [PubMed] [Google Scholar]
- 30.Sharp P.M., Bailes E., Grocock R.J., Peden J.F., Sockett R.E. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 2005;33:1141–1153. doi: 10.1093/nar/gki242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janga S.C., Moreno-Hagelsieb G. Conservation of adjacency as evidence of paralogous operons. Nucleic Acids Res. 2004;32:5392–5397. doi: 10.1093/nar/gkh882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner A. Asymmetric functional divergence of duplicate genes in yeast. Mol. Biol. Evol. 2002;19:1760–1768. doi: 10.1093/oxfordjournals.molbev.a003998. [DOI] [PubMed] [Google Scholar]
- 33.Gu X., Zhang Z.Q., Huang W. Rapid evolution of expression and regulatory divergences after yeast gene duplication. Proc. Natl Acad. Sci. USA. 2005;102:707–712. doi: 10.1073/pnas.0409186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs G.H., Rackham O., Stockwell P.A., Tate W., Brown C.M. Transterm: a database of mRNAs and translational control elements. Nucleic Acids Res. 2002;30:310–311. doi: 10.1093/nar/30.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.