Abstract
Ankylosing spondylitis (AS) is a common inflammatory arthritis predominantly affecting the axial skeleton. Susceptibility to the disease is thought to be oligogenic. To identify the genes involved, we have performed a genomewide scan in 185 families containing 255 affected sibling pairs. Two-point and multipoint nonparametric linkage analysis was performed. Regions were identified showing “suggestive” or stronger linkage with the disease on chromosomes 1p, 2q, 6p, 9q, 10q, 16q, and 19q. The MHC locus was identified as encoding the greatest component of susceptibility, with an overall LOD score of 15.6. The strongest non-MHC linkage lies on chromosome 16q (overall LOD score 4.7). These results strongly support the presence of non-MHC genetic-susceptibility factors in AS and point to their likely locations.
Introduction
Ankylosing spondylitis (AS) is the second-most-common cause of inflammatory arthritis worldwide, with a prevalence of 1/1,000–3/1,000 in white populations (Calin 1998). It is characterized by inflammation in the spine and sacroiliac joints, causing initial bone and joint erosion and subsequent ankylosis. Arthritis affecting peripheral joints, particularly the hips, occurs in 40% of cases, and inflammation may also involve extraarticular sites such as the uvea, tendon insertions, aorta, lungs, and kidneys. Genetic factors were implicated in the etiology of the disease long ago, with the demonstration of high disease familiality (de Blecourt et al. 1961). The sibling recurrence-risk ratio is 82 (Brown et al. 2000b), and heritability, assessed by twin studies, is >90% (Brown et al. 1997). The recognition of the association of B27 with AS confirmed the importance of heritable factors in the disease (Brewerton et al. 1973; Schlosstein et al. 1973) and remains one of the strongest disease associations of any inflammatory human disease. In most populations that have been studied, the prevalence of AS is strongly correlated with the prevalence of the main disease-susceptibility gene, HLA-B27 (B27). Only a few families have been reported in which AS segregates independently from B27 (van der Linden et al. 1975; Gladman et al. 1986; Deshayes et al. 1987; Woodrow 1988; Brown et al. 1996; Said-Nahal et al. 2000) and only rare cases of familial B27-negative AS have been reported (Rubin et al. 1994; Skomsvoll et al. 1995), suggesting that B27 is almost essential for the inheritance of AS within families. However, only 1%–5% of B27-positive individuals develop AS, and there is increasing evidence to suggest that other genes must also be involved. B27-positive relatives of AS patients have a recurrence risk of the disease that is 5.6–16 times greater than that of B27-positive individuals in the population at large, implying the presence of non-B27 shared familial risk factors (Calin et al. 1983; van der Linden et al. 1983). Recurrence-risk modeling in AS rejects single-gene and polygenic models. Oligogenic models with between three and nine genes operating in addition to B27 fit the observed pattern of recurrence risks in relatives of patients with AS (Brown et al. 2000b). A major non-B27 contribution to susceptibility to AS is suggested by the greater concordance rate in MZ twins (63%) than in B27-positive DZ twin pairs (23%) (Brown et al. 1997).
A preliminary whole-genome screen in 105 affected-sibling-pair families with AS demonstrated strong linkage to the MHC locus (LOD 8.1) but also identified several other regions with moderate evidence of linkage (Brown et al. 1998). Six regions lying on chromosomes 2p, 2q, 3p, 10q, 11p, and 16q achieved LOD scores >1.0, with the peak non-MHC linkage occurring on chromosome 16q (LOD 2.6). Candidate-gene studies have also implicated the gene cytochrome P450 2D6, lying on chromosome 22q13.1, in susceptibility to AS (Beyeler et al. 1996; Brown et al. 2000a) and suggest the involvement of more than one MHC gene (Hohler et al. 1998; Laval et al. 2000).
Since this preliminary study, we have continued to recruit additional families (screen 2) and have reinvestigated our initial families (screen 1) with a denser marker map. The results of these screens have been analyzed both separately and as a combined cohort consisting of 185 families containing 255 affected sibling pairs (see table 1). Results of the X-chromosome mapping have been presented elsewhere (Hoyle et al. 2000).
Table 1.
Summary of Families Included in Genome Screen
|
Screen |
|||
| No. of | 1 | 2 | 1 and 2 |
| Families: | |||
| Overall | 99 | 86 | 185 |
| With two affected sibs | 87 | 69 | 156 |
| With three affected sibs | 7 | 9 | 16 |
| With four affected sibs | 2 | 1 | 3 |
| With five affected sibs | … | 1 | 1 |
| With sibling pairs in two generations | 2 | 5 | 7 |
| With a sibling trio in one generation and a sibling pair in another | … | 1 | 1 |
| With sibling trios in two generations | 1 | … | 1 |
| With other first- or second-degree relatives | 21 | 18 | 39 |
| With both parents | 50 | 29 | 79 |
| With one parent | 22 | 29 | 51 |
| With no parents | 27 | 28 | 55 |
| Sibling pairs overall | 130 | 125 | 255 |
| Affected individuals | 235 | 210 | 445 |
| Unaffected individuals | 187 | 186 | 373 |
Subjects and Methods
Families with AS
This project was approved by the Central Oxford Research Ethics Committee (approval CM95.061) and the University of Toronto Ethics Committee. AS was defined according to the modified New York diagnostic criteria (van der Linden et al. 1984). All patients had been seen by a qualified rheumatologist, and the diagnosis of AS had been confirmed. To further confirm the diagnosis, all cases either were examined or were interviewed by telephone by one of us (L.B., M.A.B., or L.R.). In patients with atypical histories or for whom radiographs had not been performed previously, pelvic and lumbosacral spine radiographs were obtained, and attending physicians were contacted to confirm the diagnosis. All family members were typed for HLA-B27 (see below), and all the affected individuals were B27-positive. All family members were of white descent. Among participants affected with AS, the mean age at time of recruitment was 46 years (range 16–86 years), 63% were male, and 44% reported having had iritis, 16% psoriasis, and 9% inflammatory bowel disease.
Two cohorts of families were recruited (screens 1 and 2—see table 1 for family details). Six families included in our initial publication have been excluded from the current study because of changes of diagnosis or paternity (outlined below). Although both screens have similar numbers of affected sibling pairs, there are significant differences between the two cohorts. Screen 2 has fewer families (86) than screen 1 (99) and fewer affected individuals (210) than screen 1 (235). Blood samples were available from a higher proportion of parents in screen 1 families (62% of all parents) than of parents in screen 2 families (51% of all parents). Thus, it is likely that the power to detect linkage was higher for the screen 1 families than the screen 2 families.
Genotyping
All individuals were typed for HLA-B27 by PCR-SSP (Brown et al. 1996). Screen 1 families were genotyped for 505 autosomal microsatellite markers, including 259 markers from the Medical Research Council (United Kingdom) set (Reed et al. 1994), 5 markers lying within the MHC locus (62A, 82-1, 82-2, T2, and D3A) (Hsieh et al. 1997), and 241 additional markers from the Applied Biosystems Prism Linkage Mapping Set Version 2 (LMSv2) marker set (PE Biosystems). Screen 2 families were genotyped for 367 markers from the LMSv2 marker set (not including markers excluded by error-checking procedures). The markers were amplified by methods reported elsewhere (Brown et al. 1998), pooled in sets of 15–20 markers, and products were separated by electrophoresis in 6% polyacrylamide gels using ABI 373 semiautomated sequencers. Products were sized using the program GENESCAN 672, version 1.1 (PE Biosystems), and genotypes were assigned semiautomatically using the program GENOTYPER, version 1.1 (PE Biosystems).
Statistical Analysis
To minimize data errors, extensive checking procedures were employed. Mendelian inheritance of markers was checked manually within GENOTYPER, and the program GAS, version 2.0 (A. Young, unpublished), was used to convert the size data into discrete allele numbers. Consistency of allele assignment was ensured by use of a control sample on each gel and by comparison of allele distributions between screens. The program PEDCHECK (O'Connell and Weeks 1998) then was used to screen all data for previously undetected inconsistencies of Mendelian inheritance. The relationship between pedigree members was then examined, using the results of >90 markers, by means of the program SIBERR (Ehm et al. 1998). This program identifies misspecification of probable MZ twins, half-siblings, and unrelated individuals as full sibling pairs by comparison of the number of alleles shared IBD at unlinked loci with null hypothesis expectations. One previously unrecognized MZ twin pair was identified by this program and was removed from the analysis. This pair was identical at 181 of 182 alleles checked (0.5% genotyping-error rate). Excess recombination events between markers, a finding suggestive of genotyping error, was screened for, by calculation of recombination distances, using the program SIMWALK2 (Sobel and Lange 1996). Of the 527 markers used, 3 were removed from the screens as a result of this analysis. Finally, non-Mendelian errors were investigated further using the program SIBMED (Douglas et al. 2000). SIBMED identifies likely genotyping errors and marker mutations by calculating the posterior probability of an error for each sibling-pair–marker combination. The prior probability of genotype error was set at 1%, and genotypes of sibling-pair–marker combinations with a posterior probability of error of >50% were checked. Among the 614,929 genotypes scored, 51 genotype errors (0.008%) were identified by this program and were removed from the analysis.
Allele frequencies were calculated, from all scored genotypes, by means of the program DOWNFREQ (J. Terwilliger). Marker positions were obtained from public databases (either the Whitehead Institute for Biomedical Research database or GeneMap'99).
Multipoint analysis theoretically is more accurate in identifying the position of maximum linkage and has greater power to identify linkage but is susceptible to biases that are less important in two-point analysis. Multipoint analysis depends critically on the correct marker order and intermarker distances. Genotyping errors in two-point analysis affect only the marker involved, whereas, in multipoint analysis, they can also affect surrounding markers. Therefore, both analyses are presented.
Two-point nonparametric affected-sibling-pair linkage analysis was performed using the program ANALYZE (Satsangi et al. 1996). Multipoint nonparametric linkage analysis was performed using the ALL statistic of the program GENEHUNTER-PLUS (Kong et al. 1997). IBD sharing by affected sibling pairs was determined using GENEHUNTER, version 2.0 (Pratt et al. 2000), and confidence intervals for locus-specific λ values were determined by methods reported elsewhere (Cordell and Olson 1997). The contribution of each locus to the overall sibling recurrence risk was calculated assuming a sibling recurrence-risk ratio of 82 (Brown et al. 2000b) and either additive (Norman et al. 1998) or multiplicative (Risch 1987) interaction between loci. “Suggestive” and “significant” linkage are defined, according to published recommendations for affected-sibling-pair nonparametric analysis, as LOD ⩾ 2.2 and LOD ⩾ 3.6, respectively (Lander and Kruglyak 1995).
Results
Two-point results for markers achieving LOD scores of 1.0 in screens 1 and 2 and the combined results are listed in table 2. Twenty-eight markers from 14 regions achieved LOD scores of ⩾1.0 in screen 1. Among these, three markers (D6S276, D10S185, and D16S422) from three regions achieved LOD scores of ⩾1.0 in screen 2. Strong linkage with the MHC locus in this set was observed, with a peak LOD score of 6.9. Outside of the MHC locus, “significant” linkage was observed with marker D9S1826 (LOD 3.9). “Suggestive” linkage was observed for markers D10S597 (LOD 2.4) and D16S289 (LOD 2.7). In screen 2, 22 markers from 11 regions achieved LOD scores of ⩾1.0. Of these, three markers (D6S276, D10S185, and D16S422) from three regions achieved linkage with LOD 1.0 in screen 1. Strong linkage was again observed with the MHC locus (LOD 4.8). “Significant” linkage was only observed with the microsatellites in the region of the MHC locus. Outside of the MHC locus, “suggestive” linkage was observed with markers D7S519 (LOD 2.6), D19S414 (LOD 2.5), and D19S420 (LOD 3.58). Considering the combined data across both screens, 34 markers from 14 regions achieved LOD scores of ⩾1.0. Five markers from four regions achieved “suggestive” linkage (markers D1S255 [LOD 2.2], D9S288 [LOD 2.3], D9S1682 [LOD 2.3], D9S1826 [LOD 2.8], and D16S422 [LOD 3.3]).
Table 2.
Two-Point Linkage Results for Screens 1 and 2 and the Combined Data Set, Using the Program ANALYZE[Note]
|
Screen 1 |
Screen 2 |
Screens 1 and 2 |
|||||
| Chromosomeand Marker | Distance From p Telomere (cM) | LOD | P | LOD | P | LOD | P |
| 1: | |||||||
| D1S199 | 47.7 | 1.2 | .01 | .0 | .41 | .5 | .07 |
| D1S255 | 66.6 | 1.7 | .0029 | .6 | .046 | 2.2 | .0007 |
| D1S197 | 78.3 | 1.0 | .014 | .0 | .5 | .5 | .074 |
| D1S484 | 173.9 | 1.0 | .016 | .4 | .083 | 1.4 | .0053 |
| D1S2836 | 290.1 | .6 | .045 | .7 | .032 | 1.3 | .007 |
| 2: | |||||||
| D2S391 | 73.8 | 1.0 | .017 | .4 | .083 | 1.3 | .0073 |
| D2S337 | 84.1 | 1.3 | .0079 | .1 | .23 | 1.1 | .014 |
| D2S160 | 127.4 | 1.1 | .011 | .2 | .18 | 1.1 | .011 |
| D2S347 | 135.7 | .7 | .034 | .4 | .076 | 1.2 | .011 |
| D2S335 | 182.5 | 1.2 | .0083 | .0 | .5 | .5 | .069 |
| D2S157 | 212.6 | 1.6 | .0034 | … | … | 1.6 | .0034 |
| 3: | |||||||
| D3S1300 | 79 | 1.1 | .012 | .0 | .5 | .4 | .092 |
| D3S1314 | 218.3 | .0 | .5 | 1.4 | .0061 | .6 | .044 |
| 5: | |||||||
| D5S400 | 174.3 | .0 | .5 | 1.4 | .0056 | .6 | .044 |
| 6: | |||||||
| D6S309 | 13.6 | .0 | .5 | 1.5 | .0043 | .8 | .026 |
| D6S470 | 17.7 | .1 | .26 | 3.1 | .000079 | 2.2 | .00066 |
| D6S289 | 29.55 | .8 | .028 | 1.9 | .0017 | 2.5 | .00033 |
| D6S422 | 35.7 | .9 | .02 | 2.9 | .00012 | 3.6 | .000023 |
| D6S276 | 44.8 | 1.8 | .0022 | 4.8 | .0000013 | 6.5 | <10−6 |
| 82II | 44.95 | 6.9 | <10−6 | … | … | 6.9 | <10−6 |
| HLADRA | 46.05 | 3.9 | .000011 | … | … | 3.9 | .000011 |
| D6S291 | 49.8 | 1.2 | .0097 | … | … | 1.2 | .0097 |
| D6S1610 | 53.9 | 1.5 | .0042 | … | … | 1.5 | .0042 |
| D6S257 | 80 | 1.0 | .015 | .7 | .033 | 1.7 | .0024 |
| D6S460 | 90 | .6 | .045 | 2.0 | .0012 | 2.4 | .00041 |
| 7: | |||||||
| D7S519 | 70.5 | .0 | .5 | 2.6 | .00025 | .6 | .045 |
| 8: | |||||||
| D8S1784 | 116.8 | .7 | .042 | .8 | .024 | 1.5 | .0043 |
| D8S514 | 128.9 | .5 | .068 | .6 | .054 | 1.0 | .014 |
| 9: | |||||||
| D9S288 | 8.8 | 1.6 | .003 | .7 | .037 | 2.3 | .00058 |
| D9S286 | 16.8 | .5 | .066 | 1.0 | .014 | 1.5 | .0044 |
| D9S161 | 50.3 | .6 | .043 | .7 | .032 | 1.4 | .006 |
| D9S283 | 93.2 | .7 | .039 | 1.2 | .01 | 1.8 | .0019 |
| D9S1682 | 132.9 | .5 | .068 | 2.1 | .001 | 2.3 | .0006 |
| D9S1826 | 160.2 | 3.9 | .000013 | .0 | .33 | 2.8 | .00016 |
| 10: | |||||||
| D10S185 | 123.3 | 1.0 | .016 | 1.1 | .012 | 2.1 | .0009 |
| D10S192 | 131.2 | 1.5 | .0041 | .2 | .2 | 1.1 | .011 |
| D10S597 | 137.6 | 2.4 | .00043 | .1 | .23 | 1.8 | .0022 |
| D10S190 | 147.2 | 1.0 | .015 | .1 | .29 | .8 | .025 |
| 11: | |||||||
| D11S922 | 3.2 | 1.1 | .014 | … | … | 1.1 | .014 |
| D11S935 | 49.6 | 1.1 | .011 | .0 | .5 | .6 | .054 |
| 15: | |||||||
| D15S165 | 20.2 | .0 | .5 | 1.6 | .003 | .4 | .078 |
| 16: | |||||||
| D16S3068 | 46.6 | 1.3 | .0068 | .0 | .5 | .0 | .39 |
| D16S515 | 90.2 | 1.8 | .0022 | .0 | .5 | 1.1 | .014 |
| D16S516 | 98.3 | .8 | .028 | .4 | .095 | 1.1 | .012 |
| D16S422 | 109.1 | 1.4 | .0049 | 1.9 | .0015 | 3.3 | .000044 |
| D16S289 | 122.1 | 2.7 | .0002 | … | … | 2.7 | .0002 |
| 17: | |||||||
| D17S831 | 6.6 | .0 | .32 | 1.2 | .0093 | 1.0 | .014 |
| 19: | |||||||
| D19S226 | 41.7 | .3 | .14 | 1.2 | .009 | 1.3 | .0065 |
| D19S414 | 53.2 | .0 | .5 | 2.5 | .00037 | .7 | .042 |
| D19S220 | 61.4 | .0 | .48 | 1.8 | .0021 | 1.1 | .013 |
| D19S420 | 66 | .0 | .44 | 3.58 | .000025 | 2.0 | .0012 |
| D19S902 | 76.2 | .4 | .096 | 1.7 | .0025 | 1.9 | .0016 |
| D19S571 | 87.7 | .3 | .13 | 1.4 | .0058 | 1.4 | .0057 |
| 21: | |||||||
| D21S266 | 49.9 | 1.1 | .011 | .4 | .092 | 1.4 | .0052 |
Note.— Markers scoring LOD ⩾1.0 in any screen are presented. Results achieving “significant” linkage (LOD ⩾3.6) are indicated in boldface italics, and those achieving “suggestive” linkage (LOD ⩾2.2 and <3.6) are underlined.
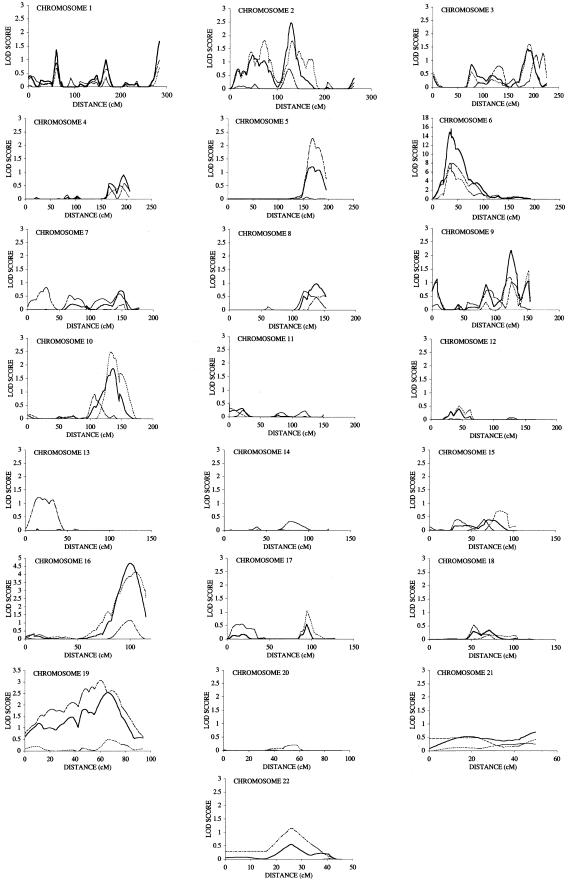
The results of the multipoint analysis are given in figure 1. For most loci, there are only minor differences between the two-point and multipoint LOD scores, but, for a small number of loci, quite marked differences were observed. For the MHC locus, the peak LOD scores obtained in screen 1, 2, and the combined screen, respectively, were 7.8, 8.1, and 15.6. The λ value for this locus overall was 5.2 (z0=0.048, z1=0.5, z2=0.45, where zn is the probability of sharing n alleles identical by descent). The contribution of the MHC locus (λ=5.2, 95% CI 3.0–9.0) to the recurrence-risk ratio in AS is either 37%, under a multiplicative disease model, or 6.8%, under an additive disease model. Multipoint analysis demonstrated greater evidence of linkage on chromosomes 2, 5, and 16 than did two-point analysis and showed less evidence of linkage on chromosomes 1 and 7. On chromosome 1, the peak multipoint linkages were at 60 cM from the p telomere, with LOD scores in screens 1 and 2 and the combined analysis being 0.6, 1.1, and 1.7, respectively. On chromosome 2, the maximum multipoint LOD scores in screens 1, 2, and the combined analysis were 1.8, 0.8, and 2.5, respectively, at 132, 125, and 132 cM from the p telomere, respectively. The magnitude of this locus is λ=1.7 (95% CI 1.3–2.3). On chromosome 5, linkage with a multipoint LOD score of 2.3 was observed at 168 cM in screen 2, compared with the two-point LOD score of 1.4. No significant evidence of linkage was observed at this locus by multipoint analysis of screen 1, resulting in a maximum LOD score, in the combined set, of only 1.2. On chromosome 7, the “suggestive” linkage observed at marker D7S519 in screen 2 was reduced in multipoint analysis to a maximum LOD score of 0.8. Two separate regions were identified on chromosome 9, although the region around D9S288 at 8.8 cM showed little support in multipoint analysis, suggesting that it may be a false-positive finding. The region around D9S283 showed “suggestive” linkage by multipoint analysis (maximum LOD score 2.2 at 124 cM), and, by two-point analysis, marker D9S1826 (at 160.2 cM) achieved a LOD score of 2.8 overall. The locus is of magnitude λ=1.5 (95% CI 1.1–2.0). A region on chromosome 10 was identified with “suggestive” linkage in screen 1 (maximum multipoint LOD score of 2.6 at 134 cM), and support in screen 2 with peak multipoint linkage of LOD 0.8 at 109 cM. In the combined data set, the maximum multipoint linkage score was LOD 1.9 at 136 cM. The overall magnitude of this locus is λ=1.2. On chromosome 16, the maximum multipoint LOD scores were greater than the two-point scores, with maximum LOD scores, for screens 1 and 2 and the combined analysis, of 4.1, 1.2, and 4.7, respectively, at 106 cM, 99 cM, and 101 cM from the p telomere, respectively. The locus magnitude is λ=1.8 (95% CI 1.3–2.4), equivalent to 13% or 2.2% of the recurrence-risk ratio for polygenic multiplicative or additive models, respectively. Multipoint analysis of chromosome 19 showed a broad region of linkage in screen 1 particularly, with peak multipoint LOD scores in screens 1 and 2 and the combined data set of 3.1, 0.5, and 2.5, at 59 cM, 65 cM and 65 cM, respectively.
Figure 1.
Multipoint results for screens 1 and 2 and the combined data set using the program GENEHUNTER PLUS. Screen 1 results are given with a dotted line, screen 2 with a dashed and dotted line, and the combined results with an unbroken line.
As previously reported, no linkage was observed on the X chromosome (P<.05) (Hoyle et al. 2000) by either two-point or multipoint analysis.
Discussion
This study provides strong evidence supporting the existence of non-MHC genes involved in AS, and points to their likely location. Regions on chromosomes 1, 2, 6, 9, 10, 16, and 19 were identified that have either “significant” or “suggestive” linkage (Lander and Kruglyak 1995) in either screen or in the combined screens.
The power of each screen to detect linkage to genes of moderate effect is only moderate. Setting the threshold for detection of LOD ⩾ 1.0, and assuming all affected sibling pairs have parents available for genotyping and are all independent, screens 1 and 2 have 80% power to detect a locus of magnitude λ=1.9 and λ=2.1, respectively (Risch 1990). Under the same assumptions, the study is estimated to have 80% power overall to detect loci of magnitude λ=1.6. Because many parents were missing in both screens and the number of independent sibling pairs is lower than the total, the actual power of this study is lower than these estimates. However, it is clear that while the study had good power overall to detect loci with moderate effects, the power of each screen to replicate findings of the other screen was not high. Therefore, loci identified in one screen may not be identified in the other. Despite this, several regions showed sufficiently strong evidence of linkage in both screens to be unlikely to be false positives.
The strong linkage demonstrated in this study with the MHC locus is further evidence confirming the central role of HLA-B27, possibly in combination with other MHC genes, in susceptibility to AS. The magnitude of this locus (λ=5.2) is considerably greater than that of any other locus identified, which were all of magnitude λ<1.9, and confirms the central role of genes encoded at this locus in susceptibility to AS. All affected relative pairs sharing 0 haplotypes identical by descent at the MHC locus were nonetheless B27-positive (identical by state), and in no family was AS inherited independently of B27. Thus, B27 appears to be essential to the inheritance of AS within families, but there is also considerable epidemiological evidence for the existence of significant non-B27 genetic susceptibility.
The strongest linkage observed outside of the MHC locus is on chromosome 16q, where maximum linkage was observed at 101 cM from the p telomere (LOD 4.7), equivalent to a genomewide significance level of <.005 (Lander and Kruglyak 1995). Both screens showed significant support for this locus, with screen 1 achieving LOD 4.1 at 106 cM and screen 2 LOD 1.2 at 99 cM, making it quite unlikely that either represents a chance finding. The region of linkage is very broad, with the 3-LOD confidence interval extending from 84 to 114 cM, and contains numerous potential candidate genes. Further refinement of this interval by high-density association/linkage disequilibrium mapping will be required to identify the actual genes involved.
Other regions identified as showing at least “suggestive” linkage included loci on chromosomes 1, 2, 9, 10, and 19. On chromosome 1, marker D1S255 achieved “suggestive” linkage overall (LOD 2.2) with support in both screens, but the multipoint linkages were weaker (LOD 1.7 overall). On chromosome 2, a region was identified by multipoint analysis, lying 123–130 cM from the p telomere. The IL-1/IL-1 RA complex is encoded in this region, and association studies of the IL-1 RA gene previously have reported association of alleles of an IL-1 RA VNTR with AS (McGarry et al. 2000; Van der Paardt et al. 2000). On chromosomes 9q and 10q, there were substantial differences in maximum points of linkage for screens 1 and 2. On chromosome 9q, although the maximum points of linkage by multipoint analysis were similar for screens 1 and 2, by two-point analysis there were substantial differences. In screen 1, marker D9S1826 achieved significant linkage (LOD 3.9; position 160.2 cM), with no support in screen 2. In screen 2, marker D9S1682, located at 132.9 cM, achieved a LOD score of 2.1 in screen 2, but a LOD of only 0.5 in screen 1. On chromosome 10, the peak position of linkage differed in screens 1 and 2 by 25 cM. By two-point analysis, one marker lying in this region (D10S192, at 131.2 cM) had good support in both screens, achieving LOD scores of 1.1, 1.0, and 2.1 in screens 1 and 2 and in the combined data set, respectively. It is a well-recognized characteristic of linkage analyses that the maximum position of linkage may vary between genome screens (Roberts et al. 1999), and, therefore, one locus may produce the different point of maximum linkage results in this region in the two screens. Further linkage mapping on chromosomes 9 and 10 may help define better the true regions of linkage in the different data sets. The region on chromosome 19 that achieved “suggestive” linkage in screen 1 (marker D19S420; LOD 3.58) achieved only minor support in screen 2 (peak multipoint LOD score 0.5), and further studies will be required to confirm this finding.
Replication of linkages in complex genetic diseases has often proved unreliable, resulting in confusion as to which linkages are true- or false-positive findings. Reasons for this include use of insufficiently stringent thresholds for identification of loci, genetic heterogeneity, and inadequate power of replication studies. To achieve 80% power to detect a locus of magnitude λ=1.8 in a complex genetic disease (equivalent to the magnitude of the chromosome 16q locus in this screen), even at a low significance threshold of LOD 1.0, will require studying 200 affected sibling pairs (with both parents available for genotyping, using a 10-cM marker map). The low power of linkage studies to replicate small genetic effects must be considered in comparisons between the results of future genetic studies and those presented here.
No other genomewide scans have been reported in AS, but scans have been completed in both psoriasis and inflammatory bowel disease, which are clinically related to AS. AS frequently complicates inflammatory bowel disease, and subclinical bowel inflammation is present in the majority of AS cases (Mielants et al. 1991). Strong linkage of chromosome 19 similar to that observed in our screen 2 was recently identified in a genomewide screen in inflammatory bowel disease (Rioux et al. 2000). The two other main IBD linkages (IBD1, located in the pericentromeric region of chromosome 16, and IBD2, located on chromosome 12) showed no evidence of linkage with AS in this study. Genomewide linkage studies in psoriatic skin disease have identified replicated linkages on chromosomes 1cen-q21, 3q21, 4q, 6p (PSORS1, the MHC region), 17q, 19p13, and 20p (Enlund et al. 1999; Samuelsson et al. 1999; Lee et al. 2000). Of these regions, we identified strong linkage at the MHC locus and on chromosome 19, weak linkage in chromosome 1q, and no linkage on chromosomes 3q21, 4q, and 17q. The peak linkage in both psoriasis and inflammatory bowel disease lies on chromosome 19p (Lee et al. 2000; Rioux et al. 2000), but, as in our own study, the area of linkage is quite broad, and considerable overlap exists. A genomewide screen has recently been reported in families with psoriatic arthritis (Samuelsson et al. 1999). Arthritis in psoriasis is clinically heterogeneous, and only a proportion of cases suffer from a spondyloarthritis similar to AS. Nonetheless, there are some overlaps with the findings presented here. Specifically, regions on chromosomes 1q, 5q, and 15p showed evidence of linkage to both diseases, adding support to our findings.
The ultimate proof that genes other than HLA-B27 are involved in AS awaits identification of the actual genes involved. The linkage results presented in this study represent an important advance in that search. This study presents strong evidence of the involvement with AS of genes localized on chromosomes 1p, 2q, 6p, 9q, 10q, 16q, and 19q. Further linkage mapping, positional candidate, and linkage disequilibrium studies are under way to define these regions better and to identify the actual genes involved.
Acknowledgments
The authors wish to thank the members of the National Ankylosing Spondylitis Society (United Kingdom) and the study families for their support of and participation in this project. We would also like to thank the staff at the following centers who assisted in recruiting families for the study: University Hospital, Birmingham (Dr. P. Jobanputra and S. Brailsford); Addenbrookes Hospital, Cambridge (Professor H. Gaston, C. Richards); Glasgow Royal Infirmary (Professor R. Sturrock, S. Armstrong); Norfolk and Norwich Hospital (Professor D. Scott); The King's Mill Centre, Mansfield (Dr. D. Walsh); Amersham Hospital (Dr. R. Stevens); Warwick Hospital (Dr. C. Marguerie, J. Gladwin-Geoghegan); Staffordshire Rheumatology Centre (Dr. A. Hassell); Princess Margaret Hospital Swindon (Dr. E. Price); West Suffolk Hospital (Dr. A. Nicholls); and York District Hospital (Dr. M. Roman). This study was supported by the Arthritis Research Campaign (United Kingdom). S.H.L. was supported by the Oliver Bird Fund of the Nuffield Foundation. S.B. and A.C. received additional support from the Coates Foundation Trust, and the Colonel W. W. Pilkington Charitable Trust. L.A.R. was supported by the Arthritis Society of Canada and the Medical Research Council of Canada. K.A.S. was supported by the Medical Research Council of Canada.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- GeneMap’99, http://www.ncbi.nlm.nih.gov/genemap99/ (for marker positions)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AS [MIM 106300]) [PubMed]
- Whitehead Institute for Biomedical Research, http://www.genome.wi.mit.edu/ (for marker positions)
References
- Beyeler C, Armstrong M, Bird HA, Idle JR, Daly AK (1996) Relationship between genotype for the cytochrome P450 CYP2D6 and susceptibility to ankylosing spondylitis and rheumatoid arthritis. Ann Rheum Dis 55:66–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewerton DA, Hart FD, Nicholls A, Caffrey M, James DC, Sturrock RD (1973) Ankylosing spondylitis and HL-A 27. Lancet 1:904–907 [DOI] [PubMed] [Google Scholar]
- Brown MA, Edwards S, Hoyle E, Campbell S, Laval S, Daly AK, Pile KD, Calin A, Ebringer A, Weeks DE, Wordsworth BP (2000a) Polymorphisms of the CYP2D6 gene increase susceptibility to ankylosing spondylitis. Hum Mol Genet 9:1563–1566 [DOI] [PubMed] [Google Scholar]
- Brown MA, Kennedy LG, MacGregor AJ, Darke C, Duncan E, Shatford JL, Taylor A, Calin A, Wordsworth P (1997) Susceptibility to ankylosing spondylitis in twins: the role of genes, HLA, and the environment. Arthritis Rheum 40:1823–1828 [DOI] [PubMed] [Google Scholar]
- Brown MA, Laval SH, Brophy S, Calin A (2000b) Recurrence risk modelling of the genetic susceptibility to ankylosing spondylitis. Ann Rheum Dis 59:883–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA, Pile KD, Kennedy LG, Calin A, Darke C, Bell J, Wordsworth BP, Cornelis F (1996) HLA class I associations of ankylosing spondylitis in the white population in the United Kingdom. Ann Rheum Dis 55:268–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA, Pile KD, Kennedy LG, Campbell D, Andrew L, March R, Shatford JL, Weeks DE, Calin A, Wordsworth BP (1998) A genome-wide screen for susceptibility loci in ankylosing spondylitis. Arthritis Rheum 41:588–595 [DOI] [PubMed] [Google Scholar]
- Calin A (1998) Ankylosing spondylitis. In: Maddison PJ, Isenberg DA, Woo P, Glass DN (eds) Oxford textbook of rheumatology, vol. 2. Oxford, Oxford University Press, pp 1058–1070 [Google Scholar]
- Calin A, Marder A, Becks E, Burns T (1983) Genetic differences between B27 positive patients with ankylosing spondylitis and B27 positive healthy controls. Arthritis Rheum 26:1460–1464 [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Olson JM (1997) Confidence intervals for relative risk estimates from affected-sib-pair data. Genet Epidemiol 14:593–598 [DOI] [PubMed] [Google Scholar]
- de Blecourt J, Polman A, de Blecourt-Meindersma T (1961) Hereditary factors in rheumatoid arthritis and ankylosing spondylitis. Ann Rheum Dis 20:215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshayes P, Demangeon S, Cavelier B, Le Loet X, Daragon A, Ropartz C (1987) Genetic study of a family of subjects with ankylosing spondylitis. Rev Rhum Mal Osteoartic 54:171–174 [PubMed] [Google Scholar]
- Douglas JA, Boehnke M, Lange K (2000) A multipoint method for detecting genotyping errors and mutations in sibling-pair linkage data. Am J Hum Genet 66:1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehm M, Wagner M (1998) A test statistic to detect errors in sib-pair relationships. Am J Hum Genet 62:181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enlund F, Samuelsson L, Enerback C, Inerot A, Wahlstrom J, Yhr M, Torinsson A, Riley J, Swanbeck G, Martinsson T (1999) Psoriasis susceptibility locus in chromosome region 3q21 identified in patients from southwest Sweden. Eur J Hum Genet 7:783–790 [DOI] [PubMed] [Google Scholar]
- Gladman DD, Urowitz MB, Anhorn KA, Chalmers A, Mervart H (1986) Discordance between HLA-B27 and ankylosing spondylitis: a family investigation. J Rheumatol 13:129–136 [PubMed] [Google Scholar]
- Hohler T, Schaper T, Schneider PM, Meyer zum Buschenfelde KH, Marker-Hermann E (1998) Association of different tumor necrosis factor alpha promoter allele frequencies with ankylosing spondylitis in HLA-B27 positive individuals. Arthritis Rheum 41:1489–1492 [DOI] [PubMed] [Google Scholar]
- Hoyle E, Laval SH, Calin A, Wordsworth BP, Brown MA (2000) The X-chromosome and susceptibility to ankylosing spondylitis. Arthritis Rheum 43:1353–1355 [DOI] [PubMed] [Google Scholar]
- Hsieh S-L, March RE, Khanna A, Cross SJ, Campbell RD (1997) Mapping of 10 novel microsatellites in the MHC class III region: application to the study of autoimmune disease. J Rheumatol 24:220–222 [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Laval SH, Bradbury L, Darke C, Brophy S, Calin A, Brown MA (2000) The role of HLA-DR genes in ankylosing spondylitis complicating inflammatory bowel disease. Rheumatology 39:64 [Google Scholar]
- Lee Y-A, Rüschendorf F, Windemuth C, Schmitt-Egenolf M, Stadelmann A, Nürnberg G, Ständer M, Wienker TF, Reis A, Traupe H (2000) Genomewide scan in German families reveals evidence for a novel psoriasis-susceptibility locus on chromosome 19p13. Am J Hum Genet 67:1020–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry F, Neilly J, Sturrock R, Field M (2000) A polymorphism within the interleukin 1 receptor antagonist (IL-1RA) gene is associated with ankylosing spondylitis (AS). Rheumatology 39:63 [DOI] [PubMed] [Google Scholar]
- Mielants H, Veys EM, Goemaere S, Goethals K, Cuvelier C, De VM (1991) Gut inflammation in the spondyloarthropathies: clinical, radiologic, biologic and genetic features in relation to the type of histology: a prospective study. J Rheumatol 18:1542–1551 [PubMed] [Google Scholar]
- Norman RA, Tataranni PA, Pratley R, Thompson DB, Hanson RL, Prochazka M, Baier L, Ehm MG, Sakul H, Foroud T, Garvey WT, Burns D, Knowler WC, Bennett PH, Bogardus C, Ravussin E (1998) Autosomal genomic scan for loci linked to obesity and energy metabolism in Pima Indians. Am J Hum Genet 62:659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt SC, Daly MJ, Kruglyak L (2000) Exact multipoint quantitative-trait linkage analysis in pedigrees by variance components. Am J Hum Genet 66:1153–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed PW, Davies JL, Copeman JB, Bennett ST, Palmer SM, Pritchard LE, Gough SC, Kawaguchi Y, Cordell HJ, Balfour KM, et al. (1994) Chromosome-specific microsatellite sets for fluorescence-based, semi-automated genome mapping. Nat Genet 7:390–395 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, McLeod RS, Griffiths AM, Green T, Brettin TS, Stone V, Bull SB, Bitton A, Williams CN, Greenberg GR, Cohen Z, Lander ES, Hudson TJ, Siminovitch KA (2000) Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet 66:1863–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N (1987) Assessing the role of HLA-linked and unlinked determinants of disease. Am J Hum Genet 40:1–14 [PMC free article] [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet 46:229–241 [PMC free article] [PubMed] [Google Scholar]
- Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS (1999) Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet 65:876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin LA, Amos CI, Wade JA, Martin JR, Bale SJ, Little AH, Gladman DD, Bonney GE, Rubenstein JD, Siminovitch KA (1994) Investigating the genetic basis for ankylosing spondylitis: linkage studies with the major histocompatibility complex region. Arthritis Rheum 37:1212–1220 [DOI] [PubMed] [Google Scholar]
- Said-Nahal R, Miceli-Richard C, Berthelot JM, Duche A, Dernis-Labous E, Le Blevec G, Saraux A, Perdriger A, Guis S, Claudepierre P, Sibilia J, Amor B, Dougados M, Breban M (2000) The familial form of spondylarthropathy: a clinical study of 115 multiplex families. Arthritis Rheum 43:1356–1365 [DOI] [PubMed] [Google Scholar]
- Samuelsson L, Enlund F, Torinsson A, Yhr M, Inerot A, Enerback C, Wahlstrom J, Swanbeck G, Martinsson T (1999) A genome-wide search for genes predisposing to familial psoriasis by using a stratification approach. Hum Genet 105:523–529 [DOI] [PubMed] [Google Scholar]
- Satsangi J, Parkes M, Louis E, Hashimoto L, Kato N, Welsh K, Terwilliger J, Lathrop G, Bell J, Jewell D (1996) Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7, and 12. Nat Genet 14:199–202 [DOI] [PubMed] [Google Scholar]
- Schlosstein L, Terasaki PI, Bluestone R, Pearson C (1973) High association of an HL-A antigen W27, with ankylosing spondylitis. N Engl J Med 288:704–705 [DOI] [PubMed] [Google Scholar]
- Skomsvoll JF, Ostensen M, Romberg O, Anda S (1995) HLA-B27 negative ankylosing spondylitis in a father and a son. Scand J Rheumatol 24:321–322 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- van der Linden J, Cats A, Keunig J, Van Rood J, Wuisman J (1975) HL-A 27 and ankylosing spondylitis. Ann Rheum Dis Suppl 34:53 [DOI] [PubMed] [Google Scholar]
- van der Linden S, Valkenburg H, Cats A (1983) The risk of developing ankylosing spondylitis in HLA-B27 positive individuals: a family and population study. Br J Rheumatol 22:18–19 [DOI] [PubMed] [Google Scholar]
- van der Linden S, Valkenburg HA, Cats A (1984) Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum 27:361–368 [DOI] [PubMed] [Google Scholar]
- Van der Paardt M, Crusius B, Van der Horst-Bruinsma I, Garcia-Gonzalez M, Baudoin P, Kostense P, Dijkmans B, Pena A (2000) Interleukin-1B and Interleukin-1 receptor antagonist gene polymorphism in ankylosing spondylitis. Arthritis Rheum 43:S264 [DOI] [PubMed] [Google Scholar]
- Woodrow JC (1988) Genetics of the spondarthropathies. Baillieres Clin Rheumatol 2:603–622 [DOI] [PubMed] [Google Scholar]