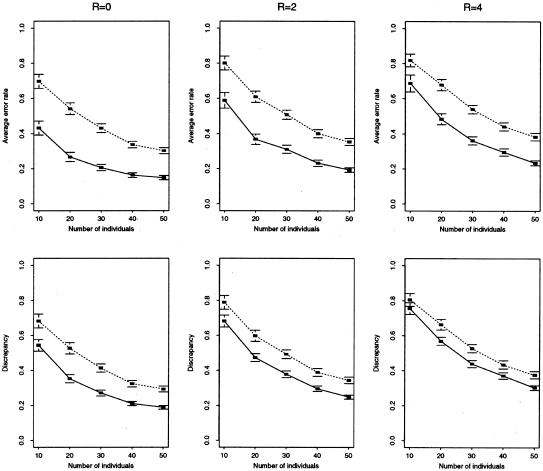
Figure 3.
Comparison of accuracy of our method (solid line) versus EM (dotted line) for microsatellite data. Top row, mean error rate (defined in text) for haplotype reconstruction. Bottom row, mean discrepancy (defined in text) for estimation of haplotype frequencies. We simulated data sets of 2n haplotypes, randomly paired to form n genotypes, for 10 equally spaced linked microsatellite loci, from a constant-sized population, under a symmetric stepwise mutation model, using a coalescent-based program kindly provided by P. N. Fearnhead. We assumed θ=4Neμ=8 (where μ is the per-generation mutation rate per locus, assumed to be constant across loci) and various values for the scaled recombination rate between neighboring loci, R=4Ner, where Ne is the effective population size and r is the genetic distance, in Morgans, between loci. For example, for humans, assuming Ne=104, and the genomewide average recombination rate, 1 cM = 1 Mb, the right-hand column would correspond to 10 kb between loci. For each combination of parameters considered, we generated 100 independent data sets. Each point thus represents an average over 100 simulated data sets. Horizontal lines above and below each point show approximate 95% confidence intervals for this average (±2 standard errors). We had difficulty getting Clark’s algorithm to consistently provide a unique haplotype reconstruction for these data.