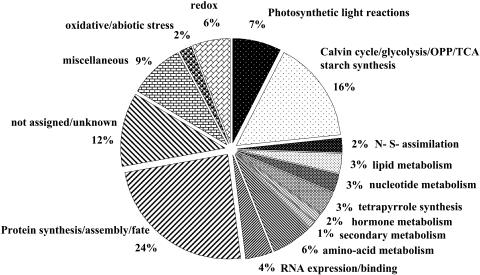
Figure 5.
Functional Summary of the 400 Identified Proteins.
To determine the functions of the identified stromal proteomes, we used in-house BLAST alignments of the identified ZmGI accessions to the predicted rice and Arabidopsis proteomes. Proteins were then functionally classified using the nonredundant MapMan functional classification system (see http://gabi.rzpd.de/projects/MapMan/) developed for Arabidopsis (Thimm et al., 2004) as a basis. BLAST alignments of proteins identified in maize resulted in a set of Arabidopsis homologs, which we classified into 25 MapMan Bins covering a wide range of pathways and functions.