Figure 1.
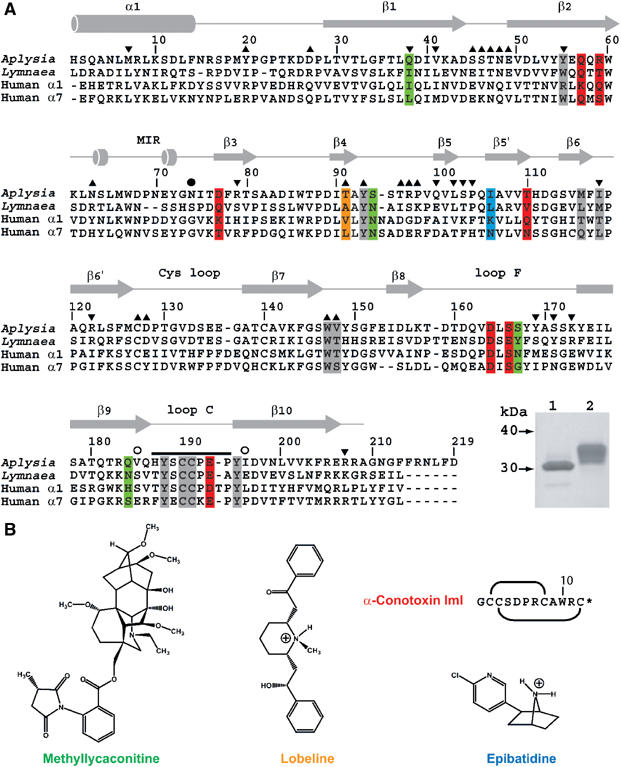
Sequence alignment of A-AChBP. (A) Structural alignment of the subunit sequences of A-AChBP (Hansen et al, 2004) and L-AChBP (Smit et al, 2001) with those of the human α1 and α7 LBDs (LGIC database). The A-AChBP sequence reported in Celie et al (2005) differs by Val substitutions at positions 43 and 138 and a N-terminal two-residue deletion. Secondary structure elements are indicated. The bar and open circles above the A-AChBP sequence indicate the loop C tip and hinge regions, respectively. The solid circle denotes the glycosylated Asn74. Tip up and down triangles denote A-AChBP residues from the (+) and (−) faces that are within a 3.5 Å radius of interaction at the subunit interface in the apo conformation. A-AChBP residues whose side chains interact within 4.5 Å with all four ligands are on a gray background. Residues specific for the nicotinic antagonists, α-conotoxin ImI and MLA, are on a red and green background and those specific for the nicotinic agonists, LOB and EPI, on an orange and a blue background, respectively. Inset: SDS–PAGE analysis (16% gel) of A-AChBP expressed from GnTI− (lane 1) and standard HEK cells (lane 2). The protein expressed in GnTI− cells migrates faster and as a thinner band, indicative of a smaller size and greater homogeneity in oligosaccharide structure. (B) Schematic view of the organic ligands, MLA (the lycoctonine ring is at the top and the N-ethylpiperidine ring at the bottom), LOB and EPI. Top right: sequence and disulfide bonding of α-conotoxin ImI; the star denotes C-terminal amidation.
