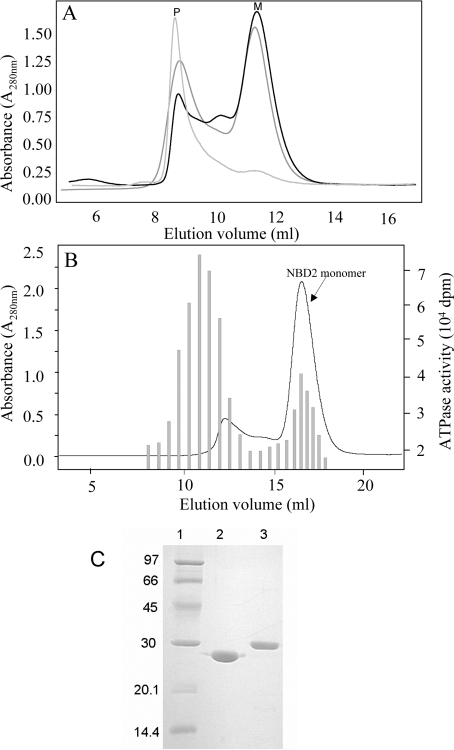
Figure 1. Purification of MRP1-NBD2.
(A) Gel-filtration chromatographic profiles of NBD2 preparations. NBD2 samples of three different preparations were applied to a gel-filtration Superdex 75 HR 10/30 column. Protein elution was followed by absorption at A280. Monomeric NBD2 was eluted in the fractions corresponding to peak M, whereas oligomers of NBD2 were recovered in the fractions corresponding to peak P. The Superdex 75 column was calibrated using the low molecular mass gel-filtration kit (Amersham Biosciences, 13.7, 25, 43, 67 and 2000 kDa) and alcohol dehydrogenase (Sigma–Aldrich, Poole, Dorset, U.K., 150 kDa). (B) ATPase activity in Superdex 200 elution fractions. An NBD2 sample purified on cobalt-affinity resin was applied to a gel-filtration Superdex 200HR 10/30 column. Protein elution was followed by A280 (line curve). ATPase activity was followed in 25 μl of aliquots of each fraction by adding 5 μl of [γ-33P]ATP (final concentration 0.3 mM, 22 Bq/pmol) to start the reaction. ATPase activity (shaded bars) was monitored by measuring the amount of [33P]Pi produced during 4 h at 30 °C. (C) SDS/PAGE of NBD1 and NBD2. Purified NBDs were analysed by SDS/PAGE (15% polyacrylamide) and revealed by Coomassie Blue staining. Lane 1, molecular mass standards; lane 2, 5 μg of NBD1; lane 3, 5 μg of NBD2.