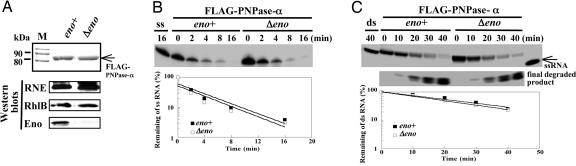
Fig. 4.
Determining the activity of PNPase-α affected by Δeno mutation. Strains containing pflag-PNP were incubated at 30°C in M9 medium plus 0.2% glycerol/40 mM succinate/1% casamino acids, then 0.05 mM isopropyl β-d-thiogalactoside was added for 2 h at OD540 = 0.6–0.1. Protein purification was as described (10). (A) Coomassie blue staining and Western blot analysis of purified FLAG–PNPase-α complexes from an isogenic pair of E. coli strains: eno wild-type [BL21(DE3), ref. 16] and eno mutant [BL21(DE3)Δeno], shown as eno+ and Δeno, respectively. The same amount of the FLAG–PNPase-α complex, with or without enolase, was used to study PNPase-α activity on single-stranded (ss) (B) and double-stranded (ds) RNA (C) substrates. “ss” indicates gel-purified 5′-32P labeled 22-mer RNA substrates incubated under the same conditions without enzyme (described in Supporting Materials and Methods); the ds-RNA unwinding reaction was carried out under the same conditions as in B, except 3 mM ATP was used. All reactions were performed at 30°C. Reaction time points are as shown. Individual reaction products in B were separated in 20% 7 M urea PAGE; the reaction products of duplex RNA in C were resolved by 16% native PAGE. Semilogarithmic plots show the remaining ss- or ds-RNA substrates revealed by the phosphorimager (FLA-5000, Fujifilm).