Table 1. Data collection and refinement statistics.
| Data set | Native | PCMBS |
|---|---|---|
| Data collection and phasing statistics | ||
| Space group | P21 | P21 |
| Cell dimensions (a, b, c) | 72.97, 87.34, 80.81 | 73.06, 87.72, 80.86 |
| Cell angles (α, β, γ), ° | 90, 111.68, 90 | 90, 112, 90 |
| Resolution, Å | 28-2.0 | 30-2.5 |
| Soaking conditions | ||
| Metal, mM | 1 | |
| Time, h | 2 | |
| Observed reflections | 217,045 | 124,310 |
| Unique reflections | 62,811 | 34,642 |
| Completeness,* % | 98.9 (94.0) | 98.9 (93.8) |
| I/σ† | 15.4 (1.6) | 32.2 (9.1) |
| Rsym‡ | 0.11 (0.59) | 0.08 (0.15) |
| Figure of merit | 0.34 | |
| Refinement | ||
| Rcrystal/Rfree | 19.2/26.6 | |
| Model composition§ | ||
| Amino acids | 951 | |
| Cofactor | FAD | FAD |
| Water molecules | 617 | |
| Ions | Cl− | |
| Total atoms | 8,137 | |
| Stereochemistry¶ | ||
| rms bond length, Å | 0.02 | |
| rms angles, ° | 2.0 | |
| Temp factors, Å2 | ||
| B-factor protein | 35.0 | |
| B-factor cofactor | 25.1 | |
| B-factor water | 43.4 |
Completeness in the highest-resolution shell (in parentheses).
I/σ in the highest-resolution shell (in parentheses).
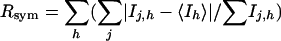 , where h is set of Miller indices and j is set of observations of reflections h.
, where h is set of Miller indices and j is set of observations of reflections h.
Model composition shows two monomers in asymmetric unit.
Over 96% of main chain dihedrals fall within the “most allowed regions” of the Ramachandran plot.
