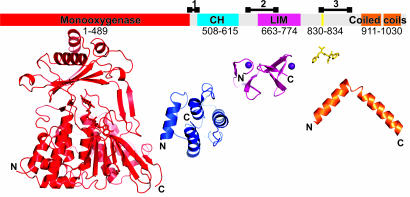
Fig. 6.
Domain organization of MICAL. In Upper, the domain structure of murine MICAL-1 is mapped onto a bar representing the linear sequence. The boundaries for the enzymatic (MO) domain (red) are taken from the mMICAL489 crystal structure. Boundaries for the CH (blue) and LIM (magenta) domains are defined from sequence alignments with homologous structures. A Src homology 3 (SH3) domain-binding motif is marked in yellow. Two coiled-coil regions are highlighted in orange, and potentially flexible linker regions are indicated by black lines above the bar (region 1, 486-512; region 2, 635-679; region 3, 830-883). Lower shows, to scale, the available structural information: CH domain, LIM domain, SH3 domain-binding motif, and examples of coiled coil (PDB ID codes 1BKR, 1IML, 1N5Z, and 1I84, respectively). Structures are orientated sequentially, with N and C termini marked.