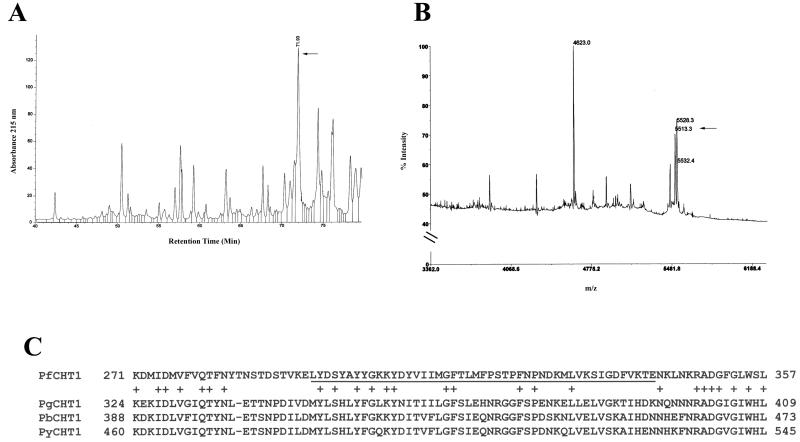
FIG. 5.
(A) Mapping of 1C3 epitope within rPfCHT1 and homology analysis. rPfCHT1 was subjected to proteolysis by Endoproteinase Glu-C and peptides separated by HPLC. (B) Dot blot immunological analysis identified one HPLC fraction as reactive with MAb 1C3 (arrow). This fraction was analyzed by Edman degradation (see the text) and matrix-assisted laser desorption ionization-time of flight MS. The ion indicated by the arrow is consistent with the single amino acid sequence identified by peptide sequencing in the fraction, taking into account the enzymatic specificity of the protease. (C) The 1C3 epitope is underlined within the amino acid stretch from 271 to 357 of the PfCHT1 coding sequence (GenBank accession no. AAF63209). Compared to homologous regions from chitinases of P. gallinaceum (PgCHT1, GenBank accession no. AAF63208), P. berghei (PbCHT1, GenBank accession no. CAC40151), and P. yoelii (PyCHT1, The Institute for Genome Research website [http://www.tigr.org/tdb/edb2/pya1/htmls/PYA1.gene.list], Locus 1275.t00001, bp 3319 to 1157, assembly c2m1275, designated “endochitinase precursor”), the 1C3 epitope has <35% identity compared to a much higher homology shared among the latter three chitinases.