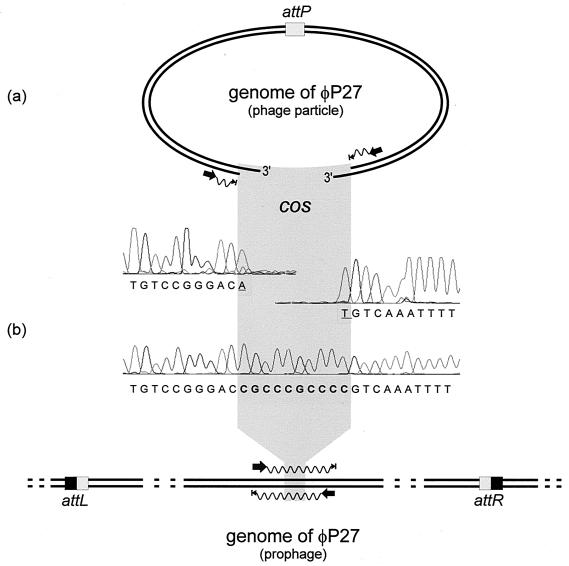
FIG. 1.
Scheme of the sequencing procedure used for the identification of cos sites in the φP27 genome. (a) Sequencing primer binding sites were selected close to the ends of the linear φP27 genome to obtain runoff sequences. Electropherograms of the runoff sequences are presented. (b) In the upper part, the DNA sequence of the corresponding region sequenced from the prophage is shown. The sequencing procedure is explained in the lower part. Black arrows indicate binding sites and the directions of primers. Waves depict cycle sequence reactions that have been performed directly from the phage DNA template (a) and from a PCR product (b). Arrowheads with bars indicate the stops of sequencing reactions. The phage attachment site (attP) and the attL and attR sites generated by insertion of the phage are highlighted by boxes. Underlined nucleotides were added by the Taq polymerase independent of the template.