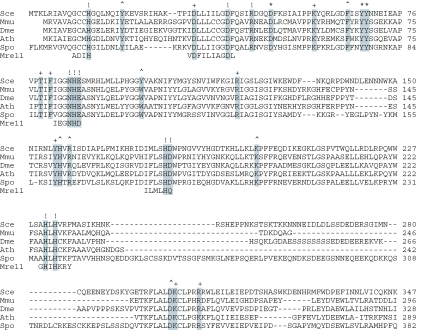
Figure 1.
Structural conservation among debranching enzymes. The amino acid sequence of S.cerevisae Dbr1 (Sce) is aligned to the homologous proteins of Mus musculus (Mmu), Drosophila melanogaster (Dme), Arabidopsis thaliana (Ath) and Schizosaccharomyces pombe (Spo) and to the metallophosphoesterase motifs of Pyrococcus furious Mre11, a DNA phosphodiesterase for which a crystal structure has been determined. The 28 conserved residues of yeast Dbr1 that were replaced by alanine in the present study are highlighted in grey. The mutational effects of alanine substitutions with respect to accumulation of branched introns are denoted above the sequence as follows: (+) indicates that there is no effect; (^) indicates 14–22% intron accumulation in vivo compared to 100% in Δdbr1 cells; (*) indicates intron accumulation between 40 and 85% of the level in Δdbr1 cells; (!) indicates intron accumulation to ≥90% of the level in Δdbr1 cells.