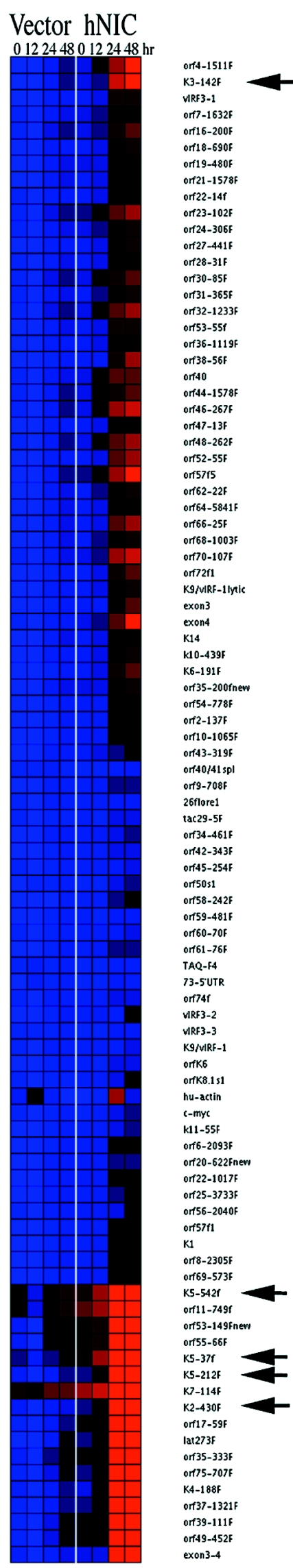
FIG.4.
Genome-wide effects of hNIC on KSHV gene expression. “Heat map” representation of KSHV mRNA upon hNIC expression was determined by real-time quantitative reverse transcription-PCR. The values were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA levels, and the medians were centered. They were clustered on the basis of a Euclidean metric and were represented on a logarithm 2 scale, to allow for a more robust statistical analysis. Black represents the decreased level and red represents the increased level relative to the median (blue) across all time points for both conditions (n = 8). Vector-expressing BCBL1 cells (Vector) and hNIC-expressing BCBL1 cells (hNIC) were examined. Arrows indicate KSHV K3, K5, and vIL-6 (K2) genes.