FIG. 1.
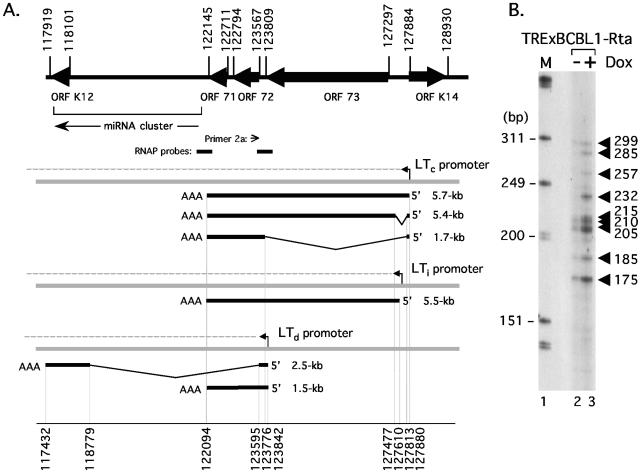
The latency gene cluster is expressed via a network of overlapping transcripts. (A) Organization of known ORFs, miRNAs, and mRNA transcripts within the major latency cluster of KSHV. Coordinates are based on the prototype BC-1 sequence (44) and correspond to initiation/termination codons (upper set) and exon sequences (lower set). A cluster of 11 miRNA genes, orientated in the direction indicated by an arrow, are located between the body of the K12 ORF (nucleotide 117,970) and the 3′ end of ORF 71 (nucleotide 121,911) (9, 40, 46). RNAP, RNase protection assay probe. A constitutively active promoter (LTc) gives rise to a precursor RNA (dotted line) that undergoes polyadenylation/cleavage and alternative splicing to produce ∼5.7-kb and ∼5.4-kb tricistronic (ORFs 71, 72, and 73) and ∼1.7-kb dicistronic (ORFs 71 and 72) mRNAs (14, 47, 50). Expression of the lytic activator RTA induces a second promoter (LTi) located between ORF 73 and LTc, giving rise to a 5.5-kb mRNA spanning all three ORFs (35). A 2.3- to 2.5-kb spliced mRNA corresponding to ORF K12, encoding multiple isoforms of kaposin, is transcribed during latency from a promoter at the 3′ end of ORF 73 and is induced further during lytic replication (27, 45). Shorter K12 transcripts initiating at 118,758 have also been reported (45). ORF K14 is essentially silent during latency but strongly induced by RTA utilizing promoter elements shared by LTi (11, 25, 30, 35). (B) Primer extension analysis to detect transcripts initiating upstream of ORF 72. Poly(A)+ RNA was isolated from mock (−Dox; lane 2) or Dox-treated (+Dox; lane 3) TRExBCBL1-Rta cells and reverse transcribed in the presence of radiolabeled oligonucleotide primer 2a. Extension products were resolved on an 8% acrylamide-7 M urea denaturing gel and visualized by autoradiography. Lane 1, size marker (in base pairs) prepared from end-labeled HinfI-digested ΦX174 DNA (Promega).