Abstract
The surface (SU) and transmembrane (TM) subunits of Moloney murine leukemia virus (Mo-MLV) Env are disulfide linked. The linking cysteine in SU is part of a conserved CXXC motif in which the other cysteine carries a free thiol. Recently, we showed that receptor binding activates its free thiol to isomerize the intersubunit disulfide bond into a disulfide within the motif instead (M. Wallin, M. Ekström and H. Garoff, EMBO J. 23:54-65, 2004). This facilitated SU dissociation and activation of TM for membrane fusion. The evidence was mainly based on the finding that alkylation of the CXXC-thiol prevented isomerization. This arrested membrane fusion, but the activity could be rescued by cleaving the intersubunit disulfide bond with dithiothreitol (DTT). Here, we demonstrate directly that receptor binding causes SU-TM disulfide bond isomerization in a subfraction of the viral Envs. The kinetics of the isomerization followed that of virus-cell membrane fusion. Arresting the fusion with lysophosphatidylcholine did not arrest isomerization, suggesting that isomerization precedes the hemifusion stage of fusion. Our earlier finding that native Env was not possible to alkylate but required isomerization induction by receptor binding intimated that alkylation trapped an intermediate form of Env. To further clarify this possibility, we analyzed the kinetics by which the alkylation-sensitive Env was generated during fusion. We found that it followed the fusion kinetics. In contrast, the release of fusion from alkylated, isomerization-blocked virus by DTT reduction of the SU-TM disulfide bond was much faster. These results suggest that the alkylation-sensitive form of Env is a true intermediate in the fusion activation pathway of Env.
The retroviruses enter cells by fusing their membranes with that of the target cell. The fusion is facilitated by the activity of the viral glycoprotein (14). The latter is composed of three copies of a two-subunit protein, Env. One of the subunits, surface (SU), has a peripheral topology, and the other, transmembrane (TM), has a transmembrane topology. The membrane fusion activity is loaded into TM but suppressed by the associated SU. The fusion is triggered when the virus binds to its cell surface receptor via SU. An exception is avian sarcoma and leukosis virus, in which complete triggering demands both receptor binding and low pH (31). The fusion activation is assumed to follow the mechanism revealed in influenza hemagglutinin (HA) (39). According to this model, the TM subunit of the retrovirus persists in a metastable state in the native Env and refolds upon receptor-induced displacement of SU. The refolding of TM involves the exposure of its N-terminal fusion peptide at the membrane-distal part of the molecule, where it can interact with the cell membrane. Further, the TM undergoes a jackknife-like backfolding. This brings the C-terminal transmembrane peptide of TM with the attached viral membrane toward the N-terminal fusion peptide in the cell membrane so that membrane fusion can take place. In the case of influenza HA, the fusion activation model has been supported by structural analyses of the ectodomains of native HA and the activated transmembrane subunit (HA2) of HA (3, 46). These showed that the three HA2 subunits formed an α-helical coiled-coil core in the native glycoprotein complex and a double-layered six-helix bundle after activation. In the latter structure, the C-terminal parts of the TM were bent as in hairpins upon the external surfaces of the centrally located N-terminal parts. In the case of the retrovirus, structural analyses of recombinant ectodomains or ectodomain peptide complexes of the TM subunit have been done (5, 11, 19, 43). These showed a six-bundle helix organization like that of the activated transmembrane subunit of influenza virus. So far, there are no reports about the native structure of the retrovirus Env, but the fact that cell entry of human immunodeficiency virus type 1 (HIV-1), human T-cell leukemia virus type 1, and avian leukosis virus (ALV) can be inhibited by peptides corresponding to the C-terminal α-helical region of the TM core (C peptide) suggests that the TM of nonactivated retrovirus also adopts a prehairpin conformation in the form of a coiled coil, which can refold into a hairpin and form the six-bundle helix (2, 5, 10, 15, 16, 45). The metastability of TM in nonactivated Env is supported by studies using ALV and Moloney murine leukemia virus (Mo-MLV), which show that fusion activating conformational changes in Env can be triggered by nonspecific protein-perturbing treatments, as in the case of influenza virus HA (40, 41).
The complete pathway of the retrovirus-mediated membrane fusion process has been mapped only insufficiently. The major reason for this is that it has been very difficult to trap intermediates in the pathway in a way that would be suitable for structural studies. Nevertheless, it has been possible to obtain important information about the pathway using suboptimal or incomplete triggering conditions. For instance, in the case of ALV, which uses both receptor binding and low pH for complete triggering of the fusion activity of Env, it has been possible to study the effects of only receptor binding on Env using a receptor ectodomain (13, 26, 27, 32). The studies indicated that receptor binding triggers an interaction between the fusion peptide of Env and the target membrane but does not lead to TM backfolding and fusion, which require low pH. In the cases of HIV-1 and simian immunodeficiency virus type 1, both of which use two receptors for triggering of fusion, it has been possible to follow the effects of the first receptor, CD 4, on the viral glycoprotein. They showed a global rearrangement of the structural domains of the peripheral gp120 subunit that facilitated binding of the second receptor and displacement from the transmembrane subunit (6, 38). Low temperature has also been used to arrest the fusion process of HIV-1 (30). Incubation at 20°C defined a post-CD 4-binding state of the glycoprotein complex, which could still be inhibited with either coreceptor interaction inhibitors or C peptides. The nature of the lipid remodeling during HIV-1 and avian sarcoma and leukosis virus fusions has been studied using lipids with positive spontaneous membrane curvature (7, 23, 30). The analyses showed that these lipids (e.g., lysophosphatidylcholine [LPC]) inhibited glycoprotein mediated cell-cell fusion. This suggests that the fusion of retrovirus involves the formation of a membrane stalk with negative surface curvature. Similar results were previously obtained for other viruses, in particular, influenza virus (8). The lipid intermediate might correspond to the hemifused state obtained at low pH with an influenza virus HA mutant carrying a glycosylphosphoinositol membrane anchor instead of the transmembrane peptide, as well as with an HA fusion peptide mutant (17, 28, 37). In this state, the outer membrane layers, but not the inner ones, have fused. The hemifused state has also been observed under neutral conditions with a Mo-MLV mutant carrying an amino acid substitution in the receptor-binding domain of Env (48). Typically, the hemifused state can resolve into complete fusion by treatment with the membrane-permeable cationic amphipath chlorpromazine (CPZ) (28). The formation of the stalk and the subsequent fusion pore might involve a concerted backfolding of TM subunits in several Env molecules at a common fusion site between the viral and the cell membranes (1, 24, 25, 30).
Recently, we demonstrated that Mo-MLV controlled its membrane fusion activity by the isomerization of its intersubunit disulfide bond (42). The SU Cys residue that participates in the bond is part of a conserved CXXC motif (34). In this motif, the other Cys residue forms a free thiol, which upon receptor binding can be activated to attack the intersubunit disulfide bond and cause its rearrangement into an alternative isomer within the motif (42). This facilitates SU dissociation and virus fusion at the cell surface. It is probable that most β-, γ-, and δ-retroviruses use this control mechanism, as they share similar CXXC-linked SU-TM disulfide bond organizations. The mechanism might have evolved to facilitate receptor-triggered cell entry at the cell surface. This is supported by the finding that the activation energy for the isomerization-controlled fusion reaction in Mo-MLV is significantly lower than for influenza virus and ALV, both of which have disulfide-linked membrane fusion protein subunits but no isomerization motif and which have to use low pH in the endosome for complete triggering of fusion (4, 20, 22, 40, 41, 44, 46).
The major evidence for the SU-TM disulfide bond isomerization-controlled fusion activation pathway of Mo-MLV was the finding that incubation of virus with receptor-positive XC cells in the presence of the membrane-impermeable alkylating agent 4-(N-maleimido)benzyl-α-trimethylammoniumiodide (M135) trapped a subfraction of the Env molecules in a CXXC-alkylated, isomerization-blocked form. The alkylated Env was fusion inactive, but the activity could be rescued by subsequent cleavage of the SU-TM disulfide bond with dithiothreitol (DTT). This suggested that receptor binding activates fusion by inducing isomerization of the SU-TM disulfide bond. However, receptor-induced isomerization was not directly demonstrated in the earlier study. In the present study, we show that receptor binding mediates isomerization of the intersubunit disulfide bond, that the disulfide rearrangement precedes the formation of a stalk of hemifused viral and cell membranes, and that the isomerization kinetically follows the virus-induced fusion. Furthermore, we demonstrate that the formation of the alkylation-sensitive form of Env follows the fusion kinetics, thus supporting its role as a true intermediate in the Env fusion activation pathway. This was corroborated by the rapid release of the fusion activity from alkylated, isomerization-blocked Env by DTT-mediated reduction of the SU-TM disulfide bond. Interestingly, alkylation-mediated fusion inhibition followed slightly faster kinetics than that of fusion. This could mean that several receptor-triggered Env molecules assemble into a common fusion site, where the alkylated form can act as a dominant inhibitor.
MATERIALS AND METHODS
Cells and virus.
XC and 3T3 cells, which were obtained from the American Type Culture Collection, Rockville, MD; MOV-3 cells, which were a gift from G. Schmidt, GSF-National Research Center for Environment and Health, Neuherberg, Germany; and Fr-57 cells (42) were maintained in Dulbecco's modified Eagle's medium (GIBCO BRL) supplemented with 10% fetal calf serum, 20 mM HEPES, and l-glutamine. Mo-MLV was prepared in MOV-3 cells and Friend MLV was prepared in Fr-57 cells by incubation for 14 h in Dulbecco's modified Eagle's medium containing 5% fetal calf serum. Radioactively labeled Mo-MLV was produced similarly but including [35S]cysteine (Amersham Pharmacia Biotech, Amersham, United Kingdom) as described previously (33, 42).
Assays.
The receptor-induced SU-TM disulfide bond isomerization was analyzed using [35S]Cys-labeled Mo-MLV (1 to 1.5 ml Mov-3 cell supernatant) that had been bound to confluent cultures of XC, 3T3, or DF-1 cells (35-mm-diameter dish) on ice for 60 min and then incubated at 37°C in fusion buffer (17 mM Tris, 8 mM HEPES, pH 7.45, 150 mM NaCl, 1.8 mM Ca2+) for the indicated times in the presence or absence of M135 (Toronto Research Chemicals Inc., North York, Canada) or lauroyl-LPC (Avanti Polar Lipids, Alabaster, AL). The virus-cell samples were then lysed in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% NP-40) for 15 min on ice in the presence of 20 mM N-ethylmaleimide (NEM) (SIGMA-Aldrich Chemie, Munich, Germany), and viral proteins were immunoprecipitated with the Mo-MLV protein-specific polyclonal antibody HE863 from Viromed Biosafety Laboratories, Camden, NJ, as described previously (33). The captured proteins were analyzed by 12% nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and quantified using a phosphorimager (33). The incubation-induced isomerization was followed by measuring the ratio of free SU to total SU, i.e., the sum of free SU and SU covalently linked to TM. As the original virus sample (medium or partially purified virus) contained significant amounts of free SU that had been released from the cells, and in addition, about 8% of the covalently linked SU-TM complexes are artificially released during SDS-PAGE sample preparation (42), we defined the free SU-to-total-SU ratio of a nonincubated sample as being representative of a nonisomerized sample. The increased ratio during incubation indicated isomerization of the SU-TM disulfide bond and was expressed as a percentage of complete isomerization. Standard deviations were calculated from four experiments. To get enough signal, we usually pooled lysates from three to six dishes for one analysis. The LPC was added directly to the culture supernatant using a 0.5 M stock solution in chloroform.
Receptor-triggered Env was trapped in its alkylation-sensitive form by incubating cell-bound [35S]Cys-labeled Mo-MLV in fusion buffer for different times at 37°C in the presence of 1.2 mM M135. In one experiment, the alkylation-incubation was followed by a short (2-min) incubation at 37°C in fusion buffer containing 20 mM DTT. After the cell cultures were washed six times with phosphate-buffered saline containing Ca2+ and Mg2+, they were lysed first for 10 min at 4°C and then for 50 min at 30°C. The alkylated, isomerization-blocked SU-TM complexes were captured by immunoprecipitation with the complex-specific monoclonal antibody (MAb) 500 (a gift from B. W. Chesebro, Rocky Mountain Laboratories, Hamilton, MT). Protein analysis and calculation of standard deviations were as described above.
We used a “fusion from without” assay to measure the efficiency by which virus fused with XC cells. This has been described elsewhere (42). Briefly, Mo-MLV was bound to confluent XC cell cultures at 0°C and then incubated at 37°C in fusion buffer for the indicated times in the presence or absence of M135, LPC, DTT, or CPZ (SIGMA-Aldrich-Chemie GmbH, Steinheim, Germany). The virus was then inactivated by treatment with pH 3 buffer (40 mM sodium citrate, pH 3, 10 mM KCl, 135 mM NaCl), and the cells were further incubated for 3 h. During the latter incubation, virus-fused cells developed into polykaryons. These were used for calculating fusion indexes and relative fusion efficiencies as described previously (42).
In some experiments, XC cells or virus was pretreated with 2 mM M135 in fusion buffer for 35 min at 37°C. Pretreatment of virus was done using ultrafiltration as described previously (42).
RESULTS
Receptor binding induces isomerization of the SU-TM disulfide bond.
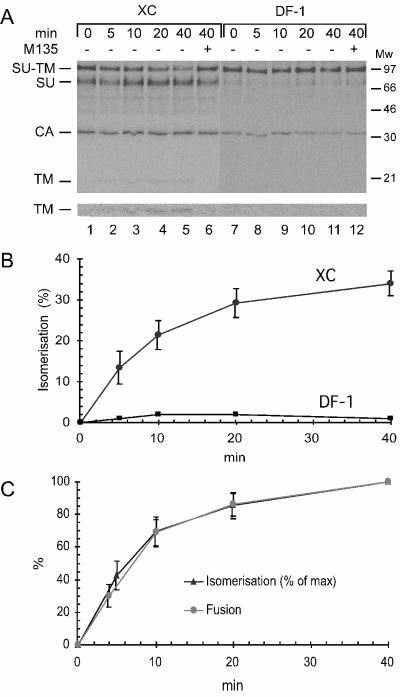
The possible receptor-induced isomerization was studied by binding [35S]Cys-labeled Mo-MLV in culture medium to receptor-positive and -negative XC and DF-1 cells and then incubating the cells in fusion buffer at 37°C for increasing times. The samples were lysed, and viral proteins were captured by immunoprecipitation for nonreducing SDS-PAGE. To follow specifically the possible isomerization in cell-bound virus, any lysis-induced isomerization of the SU-TM disulfide bond had to be blocked by including 20 mM NEM in the lysis buffer. We found that an increasing fraction of the SU-TM complexes of XC cell-bound Mo-MLV underwent isomerization with increasing time of incubation (Fig. 1A, lanes 1 to 5). The isomerization was evidenced by a decrease in SU-TM complexes, an increase in free SU, and the appearance of free TM. If the incubation of cell-bound virus was done in the presence of the membrane-impermeable alkylator M135, which modifies the CXXC thiol in receptor-triggered Envs, the isomerization reaction was interrupted and the complexes were trapped in isomerization-blocked form (Fig. 1A, lane 6). Analysis of the virus bound to the receptor-negative DF-1 cells showed that, although a corresponding amount of virus had been adsorbed to the cells by means of nonspecific interactions (35), there was a complete lack of SU-TM disulfide bond isomerization (Fig. 1A, lanes 7 to 11, and B). This was evidenced by the persistence of the SU-TM complexes. Note that free SU does not bind to DF-1 cells. Quantifications demonstrated that maximally about 35% of the SU-TM complexes in XC cell-bound virus isomerized (Fig. 1B).
FIG. 1.
(A) SU-TM disulfide bond isomerization in XC cell-bound Mo-MLV. [35S]Cys-labeled Mo-MLV in culture medium was bound to XC or DF-1 cells for 1 h on ice and incubated in fusion buffer for 0 to 40 min at 37°C. The cultures were lysed in the presence of 20 mM NEM, and viral proteins were captured by immunoprecipitation with polyclonal antibody HE863 for nonreducing SDS-PAGE. The isomerization of the SU-TM disulfide bond was followed by a decrease in SU-TM complexes, increase in free SU, and the appearance of TM. This was observed in virus bound to XC (lanes 1 to 5) but not DF-1 (lanes 7 to 11) cells. Control samples in lanes 6 (XC cell-bound virus) and 12 (DF-1 cell-bound virus) were incubated for 40 min with 1.2 mM M135. Note that free SU does not bind to the DF-1 cells, which are receptor negative. This is in contrast to virus particles, which bind nonspecifically. The figure represents a phosphorimage of the gel. The lower part shows the bottom part of the gel with the TM band at higher contrast. (B) Quantification of isomerization. The degree of isomerization was calculated based on the incubation-induced release of SU from the SU-TM complexes and expressed as a percentage of complete isomerization. In the case of virus bound to DF-1 cells, isomerization was quantified by relating the amount of SU-TM complexes to that of the nonincubated control. (C) Correlation of kinetics of isomerization with that of fusion. The isomerization kinetics was modified from panel B by setting maximal isomerization to 100%. The fusion kinetics was determined by incubating XC cell-bound Mo-MLV at 37°C in fusion buffer for 0 to 40 min and then inactivating the virus by treatment with pH 3 buffer. The cultures were further incubated for 3 h. During this time, virus-fused cells developed into polykaryons, which were used to assess relative fusion efficiencies. Maximal fusion was achieved by 40 min of incubation, which was set to 100%. Standard deviations are indicated.
To further establish the connection between SU-TM disulfide bond isomerization and fusion, we correlated the kinetics of the isomerization reaction with that of fusion. The latter was analyzed using a “fusion from without” assay with rat XC cells (42). These can, in contrast to most other cell lines, be fused with Mo-MLV (18, 36, 47). Thus, XC cell-bound virus was incubated at 37°C in fusion buffer for 0 to 40 min, the virus was inactivated with pH 3 buffer, and the efficiency of virus-induced cell-cell fusion was assessed by quantification of subsequent polykaryon formation. The analysis showed half-maximal fusion in about 7 min and maximal fusion at about 40 min (Fig. 1C), thus confirming our earlier results (42). When the fusion kinetics was correlated to that of isomerization, we found that they were very similar (Fig. 1C). We conclude that receptor binding of Env induces isomerization of the SU-TM disulfide bond and that the rearrangement kinetically follows the fusion reaction. These conclusions were further corroborated by analysis of Mo-MLV bound to receptor-positive 3T3 cells. We found that incubation at 37°C resulted in isomerization of the intersubunit disulfide bond in a significant fraction of the SU-TM complexes (data not shown). Similarly, Friend MLV underwent isomerization of the SU-TM disulfide bond in a subfraction of its Envs when incubated with XC but not DF-1 cells (data not shown).
SU-TM disulfide bond isomerization precedes virus-cell membrane hemifusion.
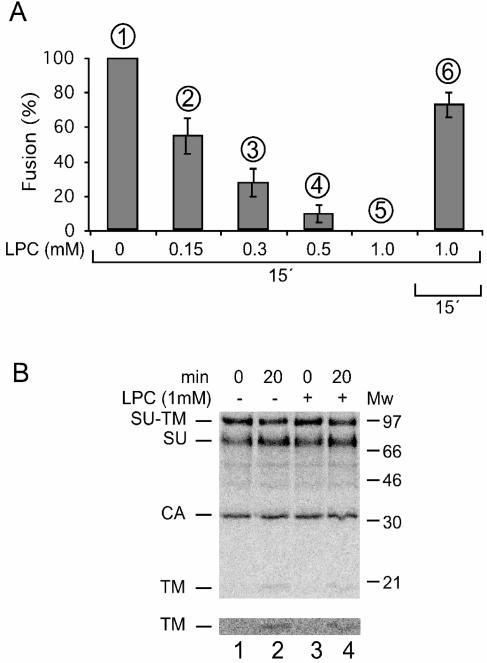
We tested whether LPC inhibited the isomerization-controlled membrane fusion of Mo-MLV. To this end, XC cell-bound Mo-MLV was incubated in fusion buffer at 37°C for 15 min in the presence of 0 to 1 mM LPC and then inactivated by pH 3 treatment. The fusion efficiency was assessed by the extent of polykaryon formation in the cell monolayer. We found that LPC inhibited virus-induced XC cell-cell fusion in a concentration-dependent manner and that 1 mM LPC inhibited fusion completely (Fig. 2A, bars 1 to 5). However, the fusion arrest was reversible. About 80% of the original fusion capacity could be rescued after the LPC-inhibited virus-cell culture was washed and reincubated in the absence of LPC (Fig. 2A, bar 6). The observed LPC-induced lipid-arrested stage suggested that Mo-MLV fusion proceeds via a stalk-like hemifusion intermediate.
FIG. 2.
(A) LPC arrests Mo-MLV fusion. Mo-MLV in culture medium was bound to XC cells and incubated in fusion buffer at 37°C for 15 min in the presence of 0 to 1 mM LPC. The virus was then inactivated by pH 3 buffer treatment. Fusion efficiencies (bars 2 to 5) relative to that of a control sample incubated in the absence of LPC (bar 1) were determined as described in the legend to Fig. 1C. Bar 6 shows the relative fusion efficiency of a parallel sample that was first incubated in 1 mM LPC and then washed four times with fusion buffer, incubated for 15 min without LPC, and finally subjected to virus inactivation treatment. Standard deviations are indicated. (B) LPC does not prevent receptor-mediated SU-TM disulfide bond isomerization. [35S]Cys-labeled Mo-MLV was bound to XC- or DF-1 cells and incubated for 0 and 20 min in fusion buffer at 37°C in the presence or absence of 1 mM LPC. Samples were processed for SU-TM disulfide bond isomerization analysis by lysis, immunoprecipitation, and SDS-PAGE as described in the legend to Fig. 1A. Note the similar degrees of SU-TM disulfide bond isomerization in lanes 2 and 4. The figure represents a phosphorimage of the gel. The lower portion shows the TM band at higher contrast.
It was important to relate the SU-TM disulfide bond isomerization event to the formation of the hemifusion intermediate. Therefore, we studied the isomerization reaction in [35S]Cys-labeled Mo-MLV that had been bound to XC cells and then incubated in the presence of 1 mM LPC. We found that the isomerization took place to the same extent in the LPC fusion-arrested virus-cell sample as in the control without LPC (Fig. 2B). This showed that the isomerization event precedes that of virus membrane-cell membrane hemifusion-stalk formation.
The accumulation of alkylated, isomerization-blocked Env during alkylation-arrested fusion correlates with fusion.
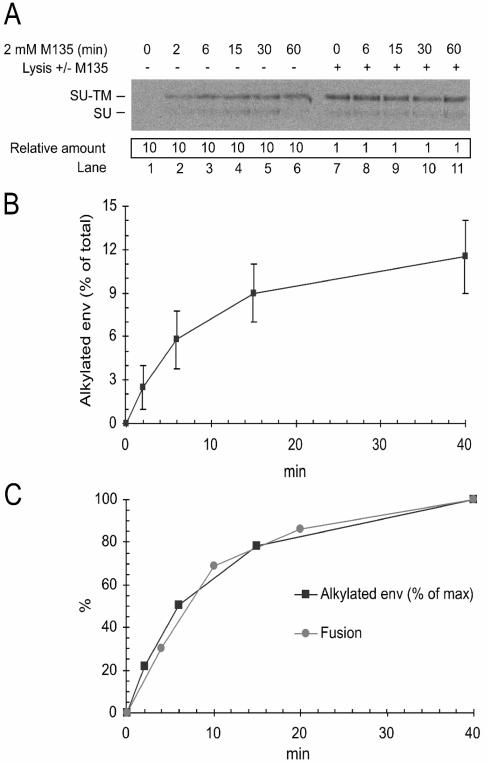
The CXXC thiol of SU cannot be alkylated in free nonactivated Mo-MLV. However when the virus is incubated with XC cells in the presence of M135, the receptor-bound fraction of Env will become alkylated and blocked in isomerization. This conditional alkylation intimates that the alkylated, isomerization-blocked Env represents a trapped intermediate in the receptor-induced fusion activation pathway. To test this possibility, we analyzed the accumulation of the alkylated, isomerization-blocked Env in time during a fusion reaction in the presence of M135. A correlation between the kinetics of accumulation of the alkylated Env and that of fusion would support a role of the alkylatable Env as an intermediate. Thus, [35S]Cys-labeled Mo-MLV was bound to XC cell cultures and incubated for 0 to 60 min at 37°C in the presence of the alkylator. The cultures were then washed, lysed, and analyzed for alkylated, isomerization-blocked SU-TM complexes by MAb 500 immunoprecipitation and subsequent nonreducing SDS-PAGE. Incubation of cell-bound virus in the presence of the alkylator converts any receptor-bound Envs to the alkylated, isomerization-blocked form, whereas the subsequent lysis in the absence of alkylator induces isomerization and subunit dissociation in the fraction of Envs that was not bound to receptors (42). Therefore, with the SU-TM complex-specific MAb 500, it is possible to follow the accumulation of receptor-bound Envs that have converted into the alkylatable form. We found that the antibody captured an increasing amount of alkylated, isomerization-blocked SU-TM complexes with increasing time of incubation (Fig. 3A, lanes 1 to 6). To find the proportion of Env in cell-bound virus that could be converted into the alkylatable fraction, we analyzed parallel samples that had been lysed in the presence of the alkylator. In this case also, the lysis-induced isomerization reaction will be aborted by the alkylation (42). Hence, MAb 500 will capture the total Env fraction. The analyses showed similar amounts of isomerization-blocked SU-TM complexes in all samples, which were significantly larger than those induced by receptor binding only (Fig. 3A, lanes 7 to 11; note differences in sample loading). Quantifications showed that the fraction of receptor-triggered alkylated Env increased to a maximum of about 12% of total Env after 40 min of incubation (Fig. 3B). The relative accumulation of alkylatable receptor-bound Env was then calculated and compared to the fusion kinetics. We found that the accumulation closely followed the fusion kinetics (Fig. 3C), thus providing biochemical support for the role of the alkylatable form of Env as a structural intermediate in the activation process of Mo-MLV Env.
FIG. 3.
(A) Accumulation of alkylated, isomerization-blocked SU-TM complexes during alkylation-inhibited fusion. XC cell-bound [35S]Mo-MLV was incubated at 37°C for 0 to 60 min in fusion buffer containing 1.2 mM M135 and then washed and lysed in the absence (lanes 1 to 6) or presence (lanes 7 to 11) of 1.2 mM M135. SU-TM complexes were captured by immunoprecipitation with the complex-specific MAb 500 and analyzed by nonreducing SDS-PAGE. Note that 10 times more sample has been applied in lanes 1 to 6 than in lanes 7 to 11. The minor amounts of SU observed in most lanes are most likely generated by artificial reduction of the SU-TM disulfide bond during sample preparation for SDS-PAGE (42). (B) Quantification of alkylated, isomerization-blocked SU-TM complexes shown in panel A, lanes 1 to 6. The relative amounts of alkylated complexes at different times of incubation were calculated as percentages of the total amount of viral Env (lanes 7 to 11). (C) Correlation of the kinetics of accumulation of the alkylated, isomerization-blocked SU-TM complexes and that of fusion. The amounts of alkylated complexes at different times of incubation were calculated as percentages of that obtained in the 40-min incubation. The fusion kinetics is from Fig. 1C.
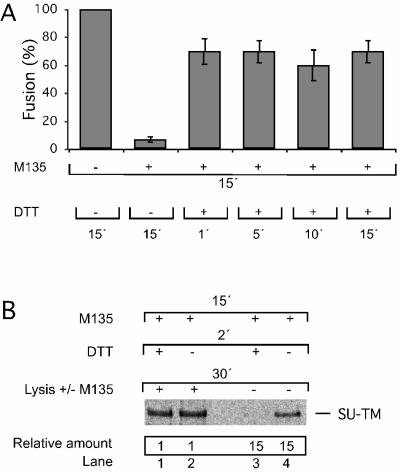
Rapid fusion after DTT cleavage of the SU-TM disulfide bond in alkylated, isomerization-blocked Envs.
The fusion kinetics of XC cell-bound Mo-MLV might reflect the time required for individual viruses to establish complete fusion sites between the virus and the cells in the culture. The fact that the alkylated, isomerization-blocked Env of receptor-bound virus accumulated, during incubation in the presence of M135, with kinetics similar to that of the fusion reaction suggested that this fraction of Env might correspond to Envs complexed with receptors in arrested fusion sites. To further test this possibility, we analyzed the fusion kinetics of Mo-MLV after release from the arrested state. We have shown that Mo-MLV can be released from this state, which we will refer to as the isomerization-arrested state (IAS), by reducing the SU-TM disulfide bond in the alkylated Envs with DTT (42). In native Env, the intersubunit disulfide bond is resistant to external reduction, but it becomes sensitive in the IAS. This restores most of the original fusion capacity of the isomerization-blocked virus. According to our model, a rapid and synchronized release of the arrested fusion activity was expected in the kinetic analysis. Thus, we bound virus to XC cell cultures and incubated them at 37°C in the presence of M135 for 15 min to chase all receptor-bound Envs into the IAS. After washing the alkylator away, we incubated the samples at 37°C in the presence of DTT for 1 to 15 min. The virus was inactivated by pH 3 treatment, and the DTT-released fusion capacity was measured by following the subsequent formation of XC cell polykaryons. The analyses showed that the shortest incubation in the presence of DTT (1 min) released the maximal fusion capacity from the IAS virus (Fig. 4A). This corresponded to about 70% of the fusion with nonarrested control virus. The reduction of the SU-TM disulfide bond in the alkylated, isomerization-blocked Env fraction of the virus was also followed using [35S]Cys-labeled Mo-MLV. In this experiment, the labeled virus was bound to XC cells and incubated first in the presence of M135 for 15 min and then in the presence of DTT for 2 min before lysis and immunoprecipitation with the complex-specific MAb 500. The lysis was done both in the absence and presence of M135. The former condition was used to specifically follow that fraction of Env that had been triggered to alkylation sensitivity by receptor binding, and the latter was used to measure the total amount of Env, i.e., including the fraction that was not triggered by receptor binding but by detergent lysis (compare the experiment shown in Fig. 3). The analyses showed that all the receptor-triggered, alkylated, and isomerization-blocked SU-TM complexes were reduced by the short DTT treatment (Fig. 4B, lanes 3 and 4). The rapid fusion release of IAS virus treated with DTT is consistent with the notion, discussed above, that IAS corresponds to virus with Env molecules trapped in arrested fusion sites.
FIG. 4.
Rapid fusion release of isomerization-arrested state. (A) Mo-MLV was bound to XC cell cultures and then incubated in fusion buffer at 37°C for 15 min in the presence of 1.2 mM M135 to accumulate it at the IAS. The alkylator was washed off, and the cultures were subjected to a second 37°C incubation in fusion buffer with 20 mM DTT for 1 to 15 min. The virus was inactivated by pH 3 treatment, and the DTT-released fusion activity was calculated relative to that of a control sample that was incubated in the absence of alkylator and DTT, as described in the legend to Fig. 1C. (B) [35S]Cys-labeled Mo-MLV was bound to XC cells and then incubated in fusion buffer at 37°C, first for 15 min in the presence of 1.2 mM M135 to induce the IAS and then for 2 min in the presence or absence of 20 mM DTT. The samples were lysed for 50 min at 30°C in the presence or absence of 1.2 mM M135. Intersubunit disulfide bond isomerization-blocked SU-TM complexes were captured by immunoprecipitation with the complex-specific MAb 500 and analyzed by nonreducing SDS-PAGE. Note that 15 times more of samples 3 and 4 than of 1 and 2 were analyzed.
IAS does not involve bilayer hemifusion.
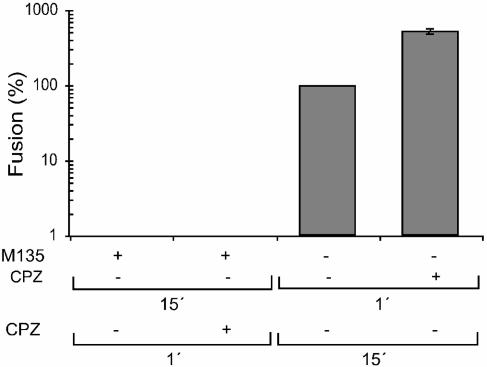
Although we showed above that isomerization of the intersubunit disulfide bond preceded the hemifusion stage of viral and cell membrane merger, it remained possible that the alkylated, isomerization-blocked Envs, which accumulated during incubation with M135, might drive the process into this stage. To find out, we probed the IAS with CPZ, which is known to resolve the hemifusion stage into complete fusion (28). The effect was compared with that of CPZ on nonarrested Mo-MLV. To this end, XC cell-bound virus was incubated first in the presence of 1.2 mM M135 for 15 min at 37°C to accumulate it at the IAS and then, after the alkylator was washed off, a second time with or without 0.4 mM CPZ for 1 min at 37°C. In parallel, virus that had been bound to XC cells was first incubated for 1 min at 37°C with or without CPZ and then for 15 min at 37°C without the drug. The virus in all samples was inactivated by low pH, and the fusion efficiencies were estimated based on subsequent polykaryon formation. The analyses showed that the virus at the IAS was not possible to drive into fusion with CPZ, whereas the drug stimulated fusion of nonarrested virus about sixfold, probably by facilitating hemifusion-to-fusion transition (Fig. 5; note the logarithmic scale). We conclude that the alkylated, isomerization-blocked Env does not induce hemifusion between the viral and the cell membranes.
FIG. 5.
Hemifusion does not occur at IAS. XC cell-bound Mo-MLV was incubated in fusion buffer at 37°C first for 15 min in the presence of 1.2 mM M135 and then for 1 min in the presence or absence of 0.4 mM CPZ. The effect of CPZ on the fusion reaction of nonarrested virus was tested by incubating cell-bound virus first for 1 min in the presence or absence of 0.4 mM CPZ and then for an additional 15 min without the drug. After the incubations, the virus was inactivated by treatment with pH 3 buffer, and the cultures were processed for polykaryon formation to assess fusion efficiencies. These were expressed as percentages of the control, i.e., the virus fused in the absence of drugs. Note the logarithmic scale.
Inhibition of fusion by alkylation is faster than fusion kinetics.
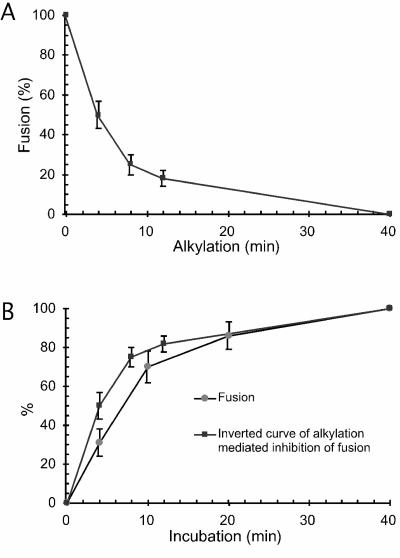
The alkylation-mediated inhibition of fusion was analyzed by subjecting XC cell-bound Mo-MLV to two-step fusion incubation for a total of 40 min. The first incubation was done for x min in the presence of M135, and the second was done for (40 − x) min in the absence of the alkylator. The fusion incubation was terminated by pH 3 buffer treatment, and the relative fusion efficiencies were estimated by the capacity of the virus to induce cell-cell fusion (polykaryons) from without. We found that the kinetics of alkylation-mediated fusion inhibition was slightly faster than the fusion kinetics (Fig. 6A and B). A trivial explanation would be that the alkylation inhibited fusion not only by blocking the CXXC thiol in the SU subunit of the virus, but also nonspecifically. However, we tested this possibility and found that pretreatment of cells or virus for 35 min at 37°C with 2 mM M135 had no detectable inhibitory effect on a subsequent fusion reaction (data not shown). Therefore, the kinetic difference could mean that the fusion activation process involves the assembly of several Env molecules into a common fusion site and that the alkylation not only blocks the fusion of sites that have been perfected during the incubation but also dominantly interferes with the activity of those incomplete ones that are completed during the subsequent incubation.
FIG. 6.
(A) Kinetics of alkylation-induced fusion inhibition. Virus, prebound to XC cells, was subjected to alkylation for 0 to 40 min by incubation at 37°C in fusion buffer containing 1.2 mM M135 and then further incubated in the absence of alkylator for a total time of 40 min. After the cells were washed with virus inactivation buffer, fused cells were allowed to rearrange into polykaryons. Fusion efficiencies relative to that of a control sample incubated for 40 min without alkylator were calculated as described in the legend to Fig. 1C. (B) Correlation between the kinetics of alkylation-induced fusion inhibition and fusion of Mo-MLV. Note that the kinetics of alkylation-induced fusion inhibition is represented as its inverse curve. The fusion kinetics is from Fig. 1C.
DISCUSSION
We showed here that a subfraction of the Envs in XC cell-bound Mo-MLV underwent SU-TM disulfide bond isomerization upon incubation at 37°C and that the kinetics of the reaction followed that of the fusion. The receptor dependence of the isomerization was evident by the fact that no rearrangement of the intersubunit disulfide bond was observed using receptor-negative chicken DF-1 cells, although these cells bound Mo-MLV as efficiently as the receptor-positive rat XC cells. The isomerization was completely blocked by alkylation of the CXXC thiol in SU. Earlier, we showed that this also arrested virus-induced cell-cell fusion but that the fusion could be rescued by reducing the SU-TM disulfide bond with DTT (42). Together, these results prove the role of the intersubunit disulfide bond isomerization in controlling the membrane fusion activity of Mo-MLV Env.
Many enveloped viruses, including the retroviruses HIV-1 and ALV, have been shown to fuse via a stalk-like membrane intermediate (23, 27, 30). This intermediate has been demonstrated by the fusion-inhibitory effect of externally added lipids like LPC, which have a strong negative spontaneous membrane curvature and therefore prevent the formation of the concave lipid surface in the stalk. We showed here that the Mo-MLV fusion was also inhibited with LPC. However, the LPC-induced lipid-arrested state did not prevent SU-TM disulfide bond isomerization, suggesting that the receptor-induced disulfide bond rearrangement precedes the remodeling of the lipid membranes. This fits a model in which the intersubunit disulfide bond isomerization triggers a membrane stalk-initiated fusion process by facilitating SU dissociation and subsequent backfolding of TM.
Earlier, we reported that the receptor-induced activation process of the membrane fusion potential in Mo-MLV Env via SU-TM disulfide bond isomerization can be blocked by alkylating the CXXC thiol (42). However, blocking required that the alkylator be present during the activation induction. Pretreatment had no effect. This suggested that the alkylatable form of Env did not represent the native structure but an intermediate in its activation pathway. Here, we tested this possibility biochemically by monitoring in time the accumulation of isomerization-blocked Env during an alkylation-aborted fusion reaction. We found that the alkylation sensitivity of Env closely followed the kinetics of fusion, as expected for a receptor-induced intermediate in the fusion activation pathway of Env.
According to the prevailing model for viral membrane fusion reaction, several fusion-activating proteins have to be collected at a fusion site between the viral and the cell membranes before membrane merger is possible. This is supported by studies of pore formation between effector and target cells that synthesize viral fusion proteins, including those of HIV-1, Mo-MLV, and ALV, and corresponding receptors (1, 23, 24, 27, 29, 30). The size of the pores suggested that several fusion proteins organize one pore. Similar conclusions have been made based on the dependence between the cell-cell fusion kinetics and the surface densities of viral fusion proteins (9). It is possible that the fusion kinetics that we observed when Mo-MLV was used to mediate XC cell-cell fusion from without (i.e., cell-virus-cell fusion) reflected differences in the time it takes for individual viruses to assemble Env-receptor complexes into fusion sites that can transform into virus-cell membrane pores similar to those observed between the effector and the target cells. Indeed cryoelectron microscopic analysis of Mo-MLV has demonstrated a large variation in the surface densities of Envs in individual particles (12). This could account for differences in the assembly kinetics of the fusion sites in different particles mediating cell-virus-cell fusion events, i.e., viruses with low Env density will take more time to complete the assembly of their fusion sites than viruses with high Env density. An interesting question, then, is whether the Envs first become activated in the putative incomplete sites or when the complete site is formed. The latter condition offers the possibility of a synchronized release of the conformational energies of several membrane fusion proteins, which has been suggested to be important for a successful merger of the viral and the cell membranes (21, 25). However, it also requires an additional triggering factor, which is dependent on the assembly of several Env-receptor complexes at a fusion site. The fact that the kinetics of the alkylation-mediated fusion inhibition was slightly faster than the fusion kinetics (Fig. 3) could mean that fusion sites composed of several Env molecules are used and that Env activation occurs even in incompletely assembled ones. Thus, according to this model, activation follows receptor binding, but final hairpin formation in TM (and subsequent membrane merger) will be successful only when several activated Env molecules come together. Consequently, when the incubation is done in the presence of M135, alkylation of the SU CXXC thiol would lead to inactivation of not only the complete but also all the incompletely assembled sites. In this case, one would expect that the fusion inhibition with M135 should show a dominant-negative effect and thus faster kinetics than the fusion. This interpretation appears to be contradicted by the findings that the kinetics of the isomerization and the accumulation of the alkylated Env so closely followed that of fusion. However, as these assays are biochemical, not functional, they lack the dimension of dominant-negative effects. Hence, minor differences in kinetics might exist, but they are not detected by our assays.
It is possible that the alkylated, isomerization-blocked Envs that are formed during Mo-MLV incubation with XC cells in the presence of M135 assemble into complete but arrested fusion sites. This notion is supported by the very rapid and efficient release of the fusion activity from the alkylated, isomerization-blocked Envs upon DTT-mediated reduction of the SU-TM disulfide bond. We have designated this state the IAS. Although fusion release from the IAS was very efficient, it did not involve bilayer hemifusion between the virus and the cells. This was shown by the complete lack of CPZ-induced fusion. Apparently, the isomerization block efficiently prevented TM hairpin formation and subsequent bilayer merger.
An inspection of the maximal level of viral Envs that underwent modification with alkylator or was isomerized in XC cell-bound virus during incubation in the presence or absence of M135 reveals an apparent dichotomy: only about 12% was alkylated, whereas as much as about 35% was isomerized. One possible explanation is that only one SU subunit in the trimeric Env complex binds to a receptor molecule at the fusion site and that the receptor-triggered activation of this SU is transmitted to the others. If receptor triggering occurs in the presence of an alkylator, this will block the isomerization reaction in all SU-TM subunit pairs of receptor-bound Envs by modifying only the CXXC thiol in the receptor-bound SUs. However, other explanations are also possible. For instance, additional Envs that have not been triggered by receptor binding might be triggered nonspecifically by the remodeling events of the viral membrane that take place during the fusion reaction. Indeed, we have demonstrated before that SU-TM disulfide bond isomerization can be triggered artificially in many ways, including by heat, urea, guanidinium hydrochloride, and NP-40 treatments (41).
The intersubunit disulfide bond isomerization-controlled fusion activation in Mo-MLV appears to provide unique possibilities to study the retrovirus-cell membrane fusion process in the future. In particular, the alkylated, isomerization-blocked Env seems to open up many interesting studies. These include structural studies of the alkylation-sensitive intermediate form of Env, as well as analysis of the molecular organization of Envs and receptors in the arrested putative fusion sites. Furthermore, the DTT rescue of the activity of the arrested sites offers a novel possibility to study the downstream events in the fusion process using synchronized conditions. For instance, it will be interesting to follow the fate of the SU-receptor complexes after they have been released from the TM subunits in the fusion site by the DTT-mediated reduction of the SU-TM disulfide bonds.
Acknowledgments
We acknowledge Mathilda Sjöberg for helpful discussions and critical reading of the manuscript.
Swedish Science Foundation grant 2778 and Swedish Cancer Foundation grant 0525 to H.G. supported this work.
REFERENCES
- 1.Blumenthal, R., D. P. Sarkar, S. Durell, D. E. Howard, and S. J. Morris. 1996. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J. Cell Biol. 135:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brighty, D. W., and S. R. Jassal. 2001. The synthetic peptide P-197 inhibits human T-cell leukemia virus type 1 envelope-mediated syncytium formation by a mechanism that is independent of Hsc70. J. Virol. 75:10472-10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 4.Carr, C. M., C. Chaudhry, and P. S. Kim. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. USA 94:14306-14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 6.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 7.Chernomordik, L. V., and M. M. Kozlov. 2003. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 72:175-207. [DOI] [PubMed] [Google Scholar]
- 8.Chernomordik, L. V., E. Leikina, V. Frolov, P. Bronk, and J. Zimmerberg. 1997. An early stage of membrane fusion mediated by the low pH conformation of influenza hemagglutinin depends upon membrane lipids. J. Cell Biol. 136:81-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danieli, T., S. L. Pelletier, Y. I. Henis, and J. M. White. 1996. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J. Cell Biol. 133:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earp, L. J., S. E. Delos, R. C. Netter, P. Bates, and J. M. White. 2003. The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH. J. Virol. 77:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol 3:465-469. [DOI] [PubMed] [Google Scholar]
- 12.Forster, F., O. Medalia, N. Zauberman, W. Baumeister, and D. Fass. 2005. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl. Acad. Sci. USA 102:4729-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez, L. D., R. J. Peters, S. E. Delos, J. A. Young, D. A. Agard, and J. M. White. 1997. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J. Cell Biol. 139:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter, E. 1997. Viral entry and receptors, p. 71-119. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 15.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 16.Jinno, A., Y. Haraguchi, H. Shiraki, and H. Hoshino. 1999. Inhibition of cell-free human T-cell leukemia virus type 1 infection at a postbinding step by the synthetic peptide derived from an ectodomain of the gp21 transmembrane glycoprotein. J. Virol. 73:9683-9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemble, G. W., T. Danieli, J. M. White, Y. I. Henis, D. L. Bodian, J. Rose, and I. A. Wilson. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76:383-391. [DOI] [PubMed] [Google Scholar]
- 18.Klement, V., W. P. Rowe, J. W. Hartley, and W. E. Pugh. 1969. Mixed culture cytopathogenicity: a new test for growth of murine leukemia viruses in tissue culture. Proc. Natl. Acad. Sci. USA 63:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobe, B., R. J. Center, B. E. Kemp, and P. Poumbourios. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. USA 96:4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leamnson, R. N., and M. S. Halpern. 1976. Subunit structure of the glycoprotein complex of avian tumor virus. J. Virol. 18:956-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leikina, E., C. Ramos, I. Markovic, J. Zimmerberg, and L. V. Chernomordik. 2002. Reversible stages of the low-pH-triggered conformational change in influenza virus hemagglutinin. EMBO J. 21:5701-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda, T., K. Kawasaki, and S. Ohnishi. 1981. Interaction of influenza virus hemagglutinin with target membrane lipids is a key step in virus-induced hemolysis and fusion at pH 5.2. Proc. Natl. Acad. Sci. USA 78:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markosyan, R. M., P. Bates, F. S. Cohen, and G. B. Melikyan. 2004. A study of low pH-induced refolding of Env of avian sarcoma and leukosis virus into a six-helix bundle. Biophys. J. 87:3291-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markosyan, R. M., F. S. Cohen, and G. B. Melikyan. 2003. HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol. Biol. Cell. 14:926-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markovic, I., E. Leikina, M. Zhukovsky, J. Zimmerberg, and L. V. Chernomordik. 2001. Synchronized activation and refolding of influenza hemagglutinin in multimeric fusion machines. J. Cell Biol. 155:833-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuyama, S., S. E. Delos, and J. M. White. 2004. Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein. J. Virol. 78:8201-8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melikyan, G. B., R. J. Barnard, R. M. Markosyan, J. A. Young, and F. S. Cohen. 2004. Low pH is required for avian sarcoma and leukosis virus Env-induced hemifusion and fusion pore formation but not for pore growth. J. Virol. 78:3753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melikyan, G. B., S. A. Brener, D. C. Ok, and F. S. Cohen. 1997. Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J. Cell Biol. 136:995-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melikyan, G. B., R. M. Markosyan, S. A. Brener, Y. Rozenberg, and F. S. Cohen. 2000. Role of the cytoplasmic tail of ecotropic Moloney murine leukemia virus Env protein in fusion pore formation. J. Virol. 74:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679-689. [DOI] [PubMed] [Google Scholar]
- 32.Netter, R. C., S. M. Amberg, J. W. Balliet, M. J. Biscone, A. Vermeulen, L. J. Earp, J. M. White, and P. Bates. 2004. Heptad repeat 2-based peptides inhibit avian sarcoma and leukosis virus subgroup a infection and identify a fusion intermediate. J. Virol. 78:13430-13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Opstelten, D. J., M. Wallin, and H. Garoff. 1998. Moloney murine leukemia virus envelope protein subunits, gp70 and Pr15E, form a stable disulfide-linked complex. J. Virol. 72:6537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinter, A., R. Kopelman, Z. Li, S. C. Kayman, and D. A. Sanders. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J. Virol. 71:8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pizzato, M., S. A. Marlow, E. D. Blair, and Y. Takeuchi. 1999. Initial binding of murine leukemia virus particles to cells does not require specific Env-receptor interaction. J. Virol. 73:8599-8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Portis, J. L., F. J. McAtee, and L. H. Evans. 1985. Infectious entry of murine retroviruses into mouse cells: evidence of a postadsorption step inhibited by acidic pH. J. Virol. 55:806-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiao, H., R. T. Armstrong, G. B. Melikyan, F. S. Cohen, and J. M. White. 1999. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell. 10:2759-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sattentau, Q. J., and J. P. Moore. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 40.Smith, J. G., W. Mothes, S. C. Blacklow, and J. M. Cunningham. 2004. The mature avian leukosis virus subgroup A envelope glycoprotein is metastable, and refolding induced by the synergistic effects of receptor binding and low pH is coupled to infection. J. Virol. 78:1403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallin, M., M. Ekstrom, and H. Garoff. 2005. The fusion-controlling disulfide bond isomerase in retrovirus Env is triggered by protein destabilization. J. Virol. 79:1678-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallin, M., M. Ekstrom, and H. Garoff. 2004. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 23:54-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 44.White, J., K. Matlin, and A. Helenius. 1981. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J. Cell Biol. 89:674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 47.Zarling, D. A., and I. Keshet. 1979. Fusion activity of virions of murine leukemia virus. Virology 95:185-196. [DOI] [PubMed] [Google Scholar]
- 48.Zavorotinskaya, T., Z. Qian, J. Franks, and L. M. Albritton. 2004. A point mutation in the binding subunit of a retroviral envelope protein arrests virus entry at hemifusion. J. Virol. 78:473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]