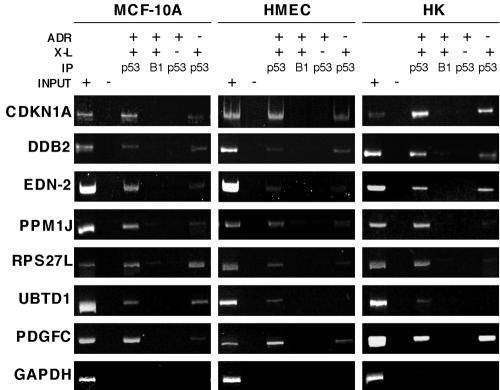
FIG. 3.
Analysis of p53 in vivo binding to consensus binding sites. Three sets each of MCF-10A cells, HMEC, and HK cells were identically processed: one set was treated with ADR (350 nM for 5 h) and formaldehyde cross-linked (ADR +, X-L +), another set was not treated with ADR and was formaldehyde cross-linked (ADR −, X-L +), and a final set was treated with ADR and not formaldehyde cross-linked (ADR +, X-L −). The DNA for PCRs was derived from p53-specific and cyclin B1-specific immunoprecipitations (IP) and amplified using primers flanking the p53 response elements in genes encoding the indicated proteins. PCRs were resolved with polyacrylamide gel electrophoresis, and the gels were stained with ethidium bromide. The cyclin B1-specific IPs were included to assess any DNA fragments purified from cross-linked lysates nonspecifically. Input +, genomic input; input −, water control. PCR results with primers directed to the coding region of GAPDH serve as a control for nonspecific DNA IP by p53-specific antibodies.