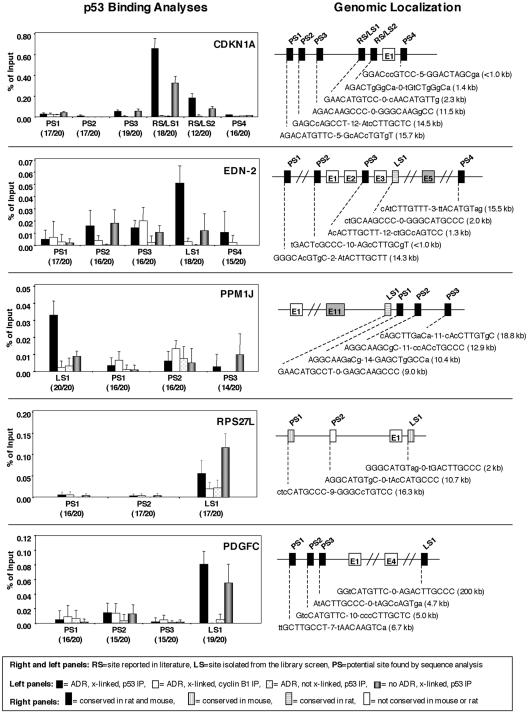
FIG. 4.
Comparative analysis of p53 binding to sites in promoter regions of known and candidate target genes. In the left panels, four sets of HMEC were processed as follows: one set was treated with ADR (350 nM for 5 h), formaldehyde cross-linked, and immunoprecipitated with a p53 antibody (solid bars); another set was treated with ADR, formaldehyde cross-linked, and immunoprecipitated with a cyclin B1 antibody (open bars); a third set was treated with ADR, not formaldehyde cross-linked, and immunoprecipitated with a p53 antibody (dotted bars); and a final set was formaldehyde cross-linked but not treated with ADR and then immunoprecipitated with a p53 antibody (gray bars). Quantitative real-time PCR was performed, and each sample was normalized to the same genomic DNA that was isolated from cells that were cross-linked and processed the same with the exception that the immunoprecipitation step was not performed. The binding sites shown are those that were recovered from the library screen (LS), those that were previously reported in the literature (RS), and those that were potential binding sites found by gene analysis using the p53MH algorithm (PS). The base pair match of the binding site to the p53 consensus is shown in parentheses. The results, shown as percentages of input DNA, are from at least three independent experiments, with the error bars representing standard deviations. The right panels show schematics of the genomic structure and localization of known and putative p53 binding sites analyzed. The bar shading indicates species conservation as indicated. Exons are indicated with an E followed by the exon number in either an open box or a shaded box (representing the terminal exon). The sequences of the binding sites present in the regions analyzed are shown, and in parentheses the distances of the binding sites from the start of exon 1 are given.