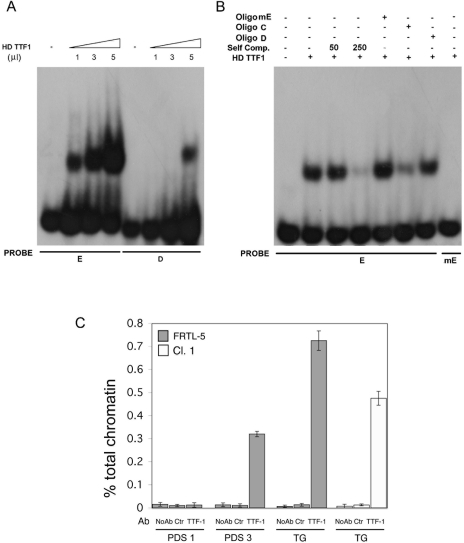
FIG. 6.
Interaction of the TTF-1 protein with the E binding site within the rat Pds promoter by EMSA and ChIP analysis on the Tg and pendrin promoter regions. (A) The recombinant TTF-1 was added to radiolabeled oligonucleotides as indicated. (B) The specificity of complexes was tested by competition analysis. A constant amount (250-fold excess or as indicated) of cold oligonucleotides was added to the reaction mixture as indicated in Materials and Methods. The mutated E probe (mE), which contains the CAAG to CGTG mutation (underlined), did not compete for the binding of E to recombinant TTF-1 protein (lane 5), nor did it bind recombinant TTF-1 protein when radiolabeled (last lane). (C) TTF-1 associated with both the rat pendrin and Tg promoters in vivo. Chromatin extracted from cross-linked FRTL-5 cells was immunoprecipitated in parallel using no antibodies (No Ab), anti-ERK (Ctr), or antibodies against TTF-1. The immunoprecipitates were analyzed by real-time PCR with oligonucleotide primers corresponding to the rat Tg promoter (TG) or rat pendrin promoter (PDS1, spanning the E site, or PDS3, spanning a distal genomic region about 15 kb upstream to the initiator ATG). Parallel PCRs were performed with total input DNAs obtain from unprecipitated aliquots of similarly treated chromatin samples. The amount of precipitated DNA was calculated relative to the total input chromatin and expressed as percentage of the total according to the formula: % total = 2ΔCt × % input chromatin, where ΔCt = Ctinput − Ctimmunoprecipitation (20). Ct is the cycle threshold within the linear range of all reactions. Standard deviations are indicated, and the experiment is representative of at least three independent experiments.