FIG. 7.
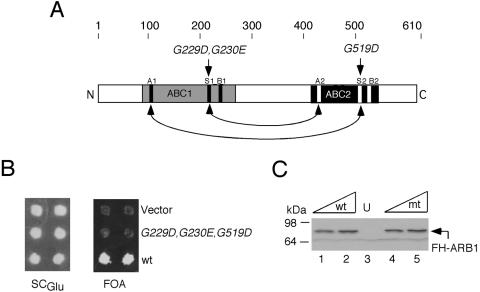
Signature sequences in the ABCs of ARB1 are essential in vivo. (A) The primary structure of ARB1 is depicted schematically with amino acid positions and the locations of mutations made in conserved residues (in italics) shown at the top, regions of sequence similarity to ATP-binding domains in other ABC proteins are shaded, and locations of the Walker A and B motifs and signature sequences (S) are indicated with black (ABC1) or white (ABC2) rectangles. The predicted interactions that would stabilize formation of a dimer between ABC1 and ABC2, sandwiching two molecules of ATP, are indicted with arrows. (B) Patches of an arb1Δ strain containing ARB1 URA3 plasmid pDH22 and LEU2 plasmids YCplac111 (empty vector; row 1), pDH144 (containing FH-ARB1-G229D-G230E-G519D; row 2), or pDH129 (containing FH-ARB1; row 3), were replica plated to SCGlu-Leu-Ura (left panel) and medium containing 5-FOA (right panel). (C) WCEs of the strains described in panel B were grown in SCGlu-Leu-Ura to an A600 of ∼1.2, and 10 or 20 μg of WCE from cells containing pDH129 (lanes 1 and 2), pDH144 (lanes 4 and 5), or empty vector (lane 3) in addition to pDH22 were subjected to Western analysis with anti-His6 antibodies. mt, mutant.