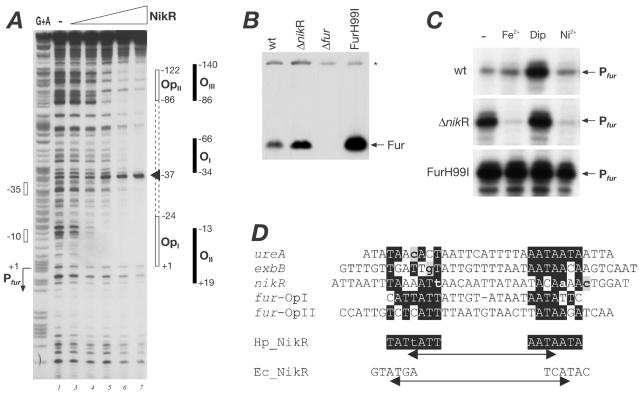
FIG. 6.
Binding and regulation of the metal-responsive regulators to the fur promoter. (A) Identification of NikR operators in the fur promoter by DNase I footprinting of NikR on a radioactively labeled Pfur probe consisting of a 220-bp BamHI-PstI fragment from plasmid pGemCl2 labeled at its BamHI site. An equivalent amount of 5′-end-labeled probe was incubated with increasing amounts of NikR, with 0, 0.011, 0.033, 0.10, 0.30, 0.9, and 2.5 μM purified protein corresponding to lanes 1 to 7, respectively. The vertical open bars on the right indicate the areas of DNase I protection resulting from binding of NikR; the filled boxes represent the positions of footprints due to the Fur protein previously reported (9, 11), and the numbers indicate the boundaries of the operators with respect to the +1 transcriptional start site of the respective promoters as indicated. Lane G+A is a G+A sequence reaction on the DNA probe used as a size marker (19). The open bars and the bent arrow on the left indicate the position of the Pfur promoter. (B) Western blot showing derepression of the Fur protein in mutants of the metal-responsive regulators NikR and Fur. Total protein extracts of the G27 wild-type strain (wt), a NikR-null mutant (ΔnikR), a Fur-null mutant (Δfur), and the mutant G27(H99I) expressing the iron-blind protein (FurH99I) collected at logarithmic phase were transferred after SDS-PAGE onto nitrocellulose and stained with anti-Fur antiserum. The band corresponding to the Fur protein is highlighted with an arrow, and the asterisk denotes a cross-reacting band which serves as a control. (C) Quantitative primer extension of the Pfur promoter showing transcriptional regulation in response to metals and metal-responsive regulators. Total RNA was extracted from cells of the G27 wild type (wt), the NikR-null mutant (ΔnikR), and G27(H99I) expressing the FurH99I iron-blind protein, which were grown to logarithmic phase (−) and then treated for 20 min. with 1 mM FeCl2 (Fe2+), 100 μM 2,2′-dipyridiyl (Dip), or 5 mM NiCl2 (Ni2+). Primer extension was performed on 10 μg of total RNA from each preparation with the Fur-R2 primer (11), and the elongation product corresponding to the Pfur transcript is indicated. (D) Alignment of the NikR operators with respect to the determinants of the urease operator. Variations in the nucleotide sequences between the available sequences are shown in lowercase gray-shaded letters. Conserved bases and the proposed consensus operator are shaded in black. The E. coli NikR consensus is reported underneath.