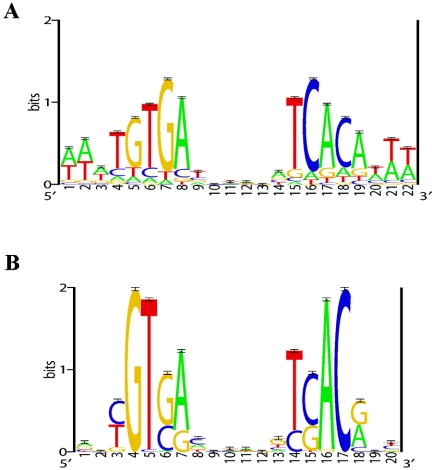
FIG. 1.
Sequence logos of the E. coli CRP motif model (A) and the putative CRPMt motif model (B). The E. coli CRP motif model represents 87 experimentally identified (DNase I footprinted) binding sites; this motif model was used as prior information in the prediction of CRPMt binding sites (see Materials and Methods). The putative CRPMt motif model represents 58 predicted sites (see Table 2) from the set of intergenic regions from the M. tuberculosis genome. Sequence logos depict the relative frequency of each base at each position of the motif. The y axis indicates the information content measured in bits, and error bars represent standard deviations at each position due to the limited sample size (58).