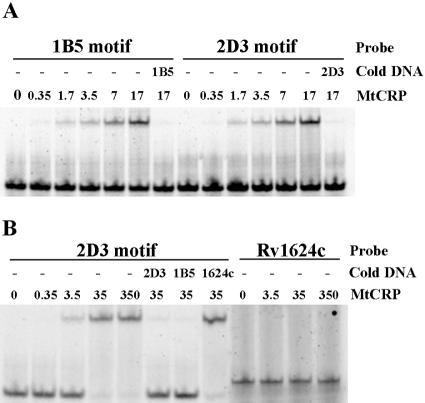
FIG. 2.
EMSA experiment showing binding of CRPMt with motifs identified in captured DNA sequences. (A) The mobility of 28-bp synthesized probes containing motifs of 1B5 and 2D3 fragments was retarded by CRPMt, and the free probes reappeared in the presence of a 500-fold excess of unlabeled probe DNA, as specified. (B) The shift of the 2D3 28-bp motif probe by CRPMt could be competed by either 2D3 or 1B5 unlabeled motif DNA, but not by the 40-bp intergenic DNA upstream of Rv1624c (1624c) that was used as a negative control. This control Rv1624c DNA probe also failed to bind to CRPMt. The labeled DNA probe, unlabeled competitor DNA (cold DNA), and amount of CRPMt (nM) that was used are specified for each lane.