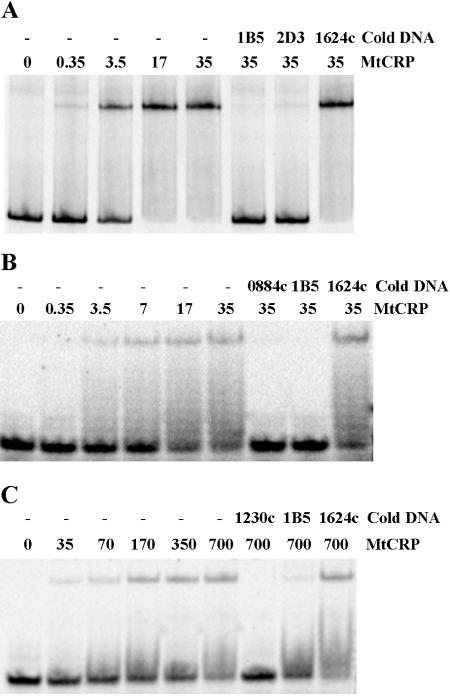
FIG. 4.
EMSA showing CRPMt interactions with representative DNA binding sites identified by Gibbs sampling. DNA probes are as follows: A, the full-length Rv0884c-Rv0885 intergenic region, which contains a motif with a high probability of belonging to the CRPMt motif model; B, the 20-bp predicted binding site in the Rv0884c-Rv0885 intergenic region; and C, the 20-bp predicted binding site upstream of the Rv1230c open reading frame, which has a low probability of belonging to the motif model. For each lane, the CRPMt concentration (nM) is noted, and “cold DNA” specifies unlabeled competitor DNA fragments used at a 500-fold excess relative to the labeled probe DNA. The 40-bp Rv1624c nonspecific DNA was used as a negative control throughout.