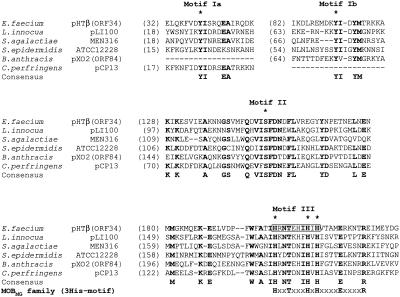
FIG. 5.
Comparison of the N-terminal region of the deduced ORF34 protein (TraI) of pHTβ with the hypothetical proteins found in sequence databases. The bold characters indicate the conserved amino acid residues in each protein. The asterisks on the sequences show the key residues, Tyr, Ser, and His3 (3His) in motifs I, II, and III, respectively. There are two motif I candidates (Ia and Ib) in most of the proteins. The gray box in motif III indicates the deleted 10-amino-acid residues of ORF34 resulting in pHTβ/ORF34del. The lowest sequence shows the putative consensus His3 motif (motif III) of these proteins (designated as MOBMG family), H(x2)T(x3)HxH(x4)E(x4)R. The accession numbers for the proteins are as follows: NP_569166 for Listeria innocua pLI100 (81,905 bp), NP_734852 for Streptococcus agalactiae MEN316, NP_765038 for Staphylococcus epidermidis ATCC 12228, NP_053238 for B. anthracis pXO2-84, and NP_150032 for Clostridium perfringens strain 13 plasmid pCP13 (54,310 bp), respectively.