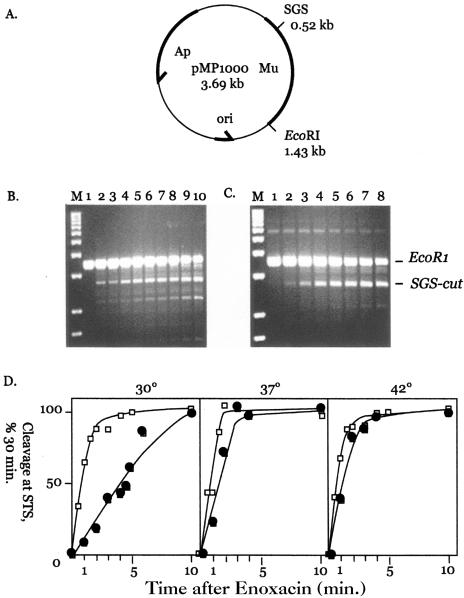
FIG. 2.
A. Physical map of plasmid pMP1000. The positions of the phage Mu DNA (bold line) and the SGS are indicated along with the position of a single EcoRI restriction site. B. Kinetics of WT gyrase cleavage in the presence of enoxacin. Plasmid pMP1000 was incubated with EcoRI and reconstituted WT gyrase as indicated at 37°C for 1 h. After the addition of enoxacin, aliquots were withdrawn and quenched with SDS and proteinase K at different times. The marker in lane M is a 1-kb DNA ladder; gyrase cleavage at the SGS produces a 2.7-kb band. Incubation with enoxacin was as follows: 0 min, lane 1; 0.5 min, lane 2; 1 min, lane 3; 1.5 min, lane 4; 2 min, lane 5; 3 min, lane 6; 4 min, lane 7; 5 min, lane 8; 10 min, lane 9; and 30 min, lane 10. C. Kinetics of GyrB652 gyrase cleavage in the presence of enoxacin. pMP1000 DNA was incubated as described above with GyrB652 gyrase. Enoxacin was added and incubation continued at 37°C for the following times: 0 min, lane 1; 1 min, lane 2; 2 min, lane 3; 3 min, lane 4; 4 min, lane 5; 5 min, lane 6; 10 min, lane 7; and 30 min, lane 8. The positions of EcoRI-linearized pMP1000 and gyrase-cleaved product are marked as EcoRI and SGS-cut at the right edge of panel C. D. Temperature effects on the kinetics of enoxacin-dependent cleavage. Cleavage reactions were carried out using WT gyrase (open squares) and GyrB652 gyrase (filled squares) at 30°C (left panel), 37°C (central panel), and 42°C (right panel). Cleavage levels were quantitated using Bio-Rad Quantity One video imaging software, and the level is given as a percentage of product formed after 30 min of incubation with enoxacin (see panels B and C). The estimated time to achieve half-maximal cleavage was about 1 min at 30°C, 45 s at 37°C, and 30 s at 42°C for WT gyrase and 5 min at 30°C, 1.6 min at 37°C, and 1 min at 42°C for GyrB652 gyrase.