Abstract
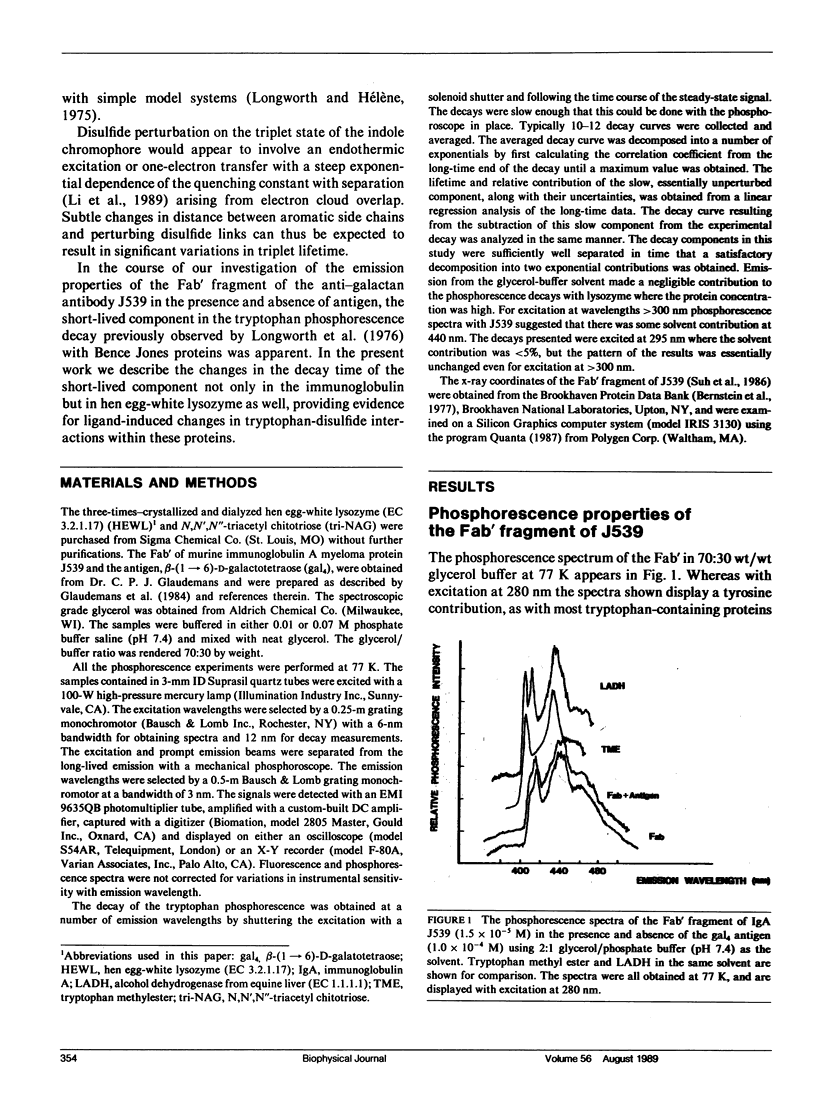
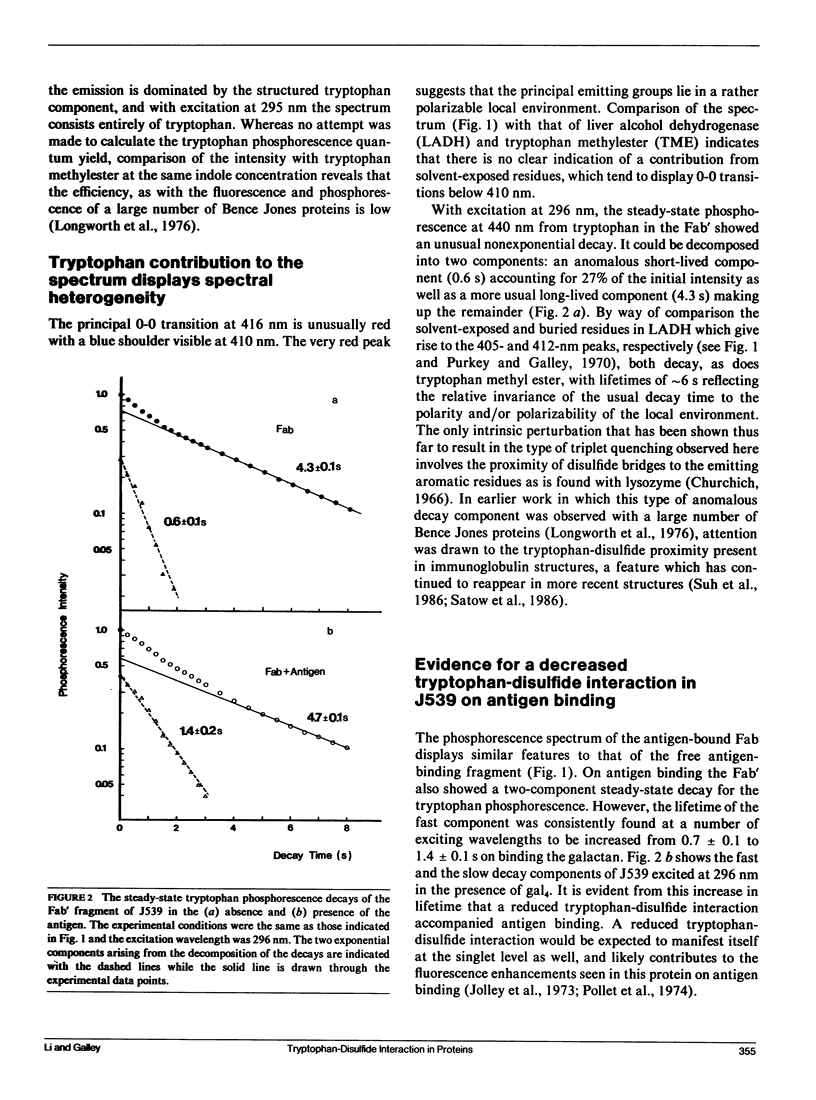
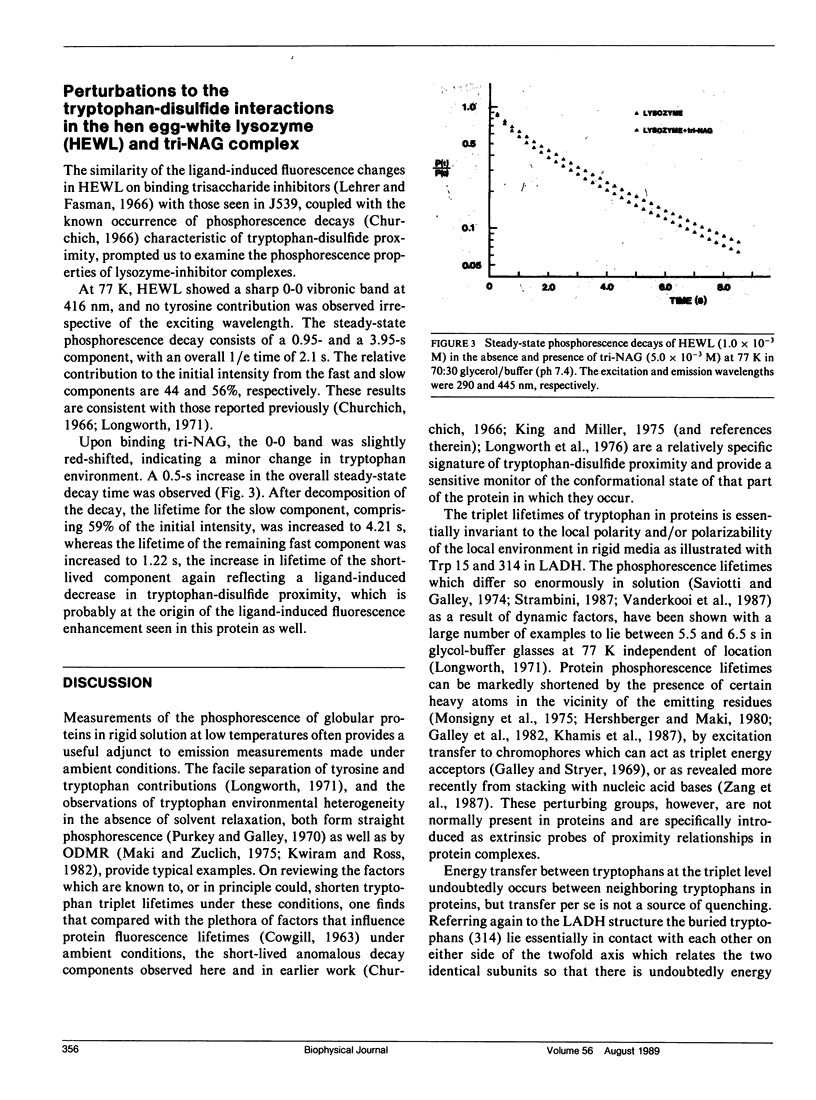
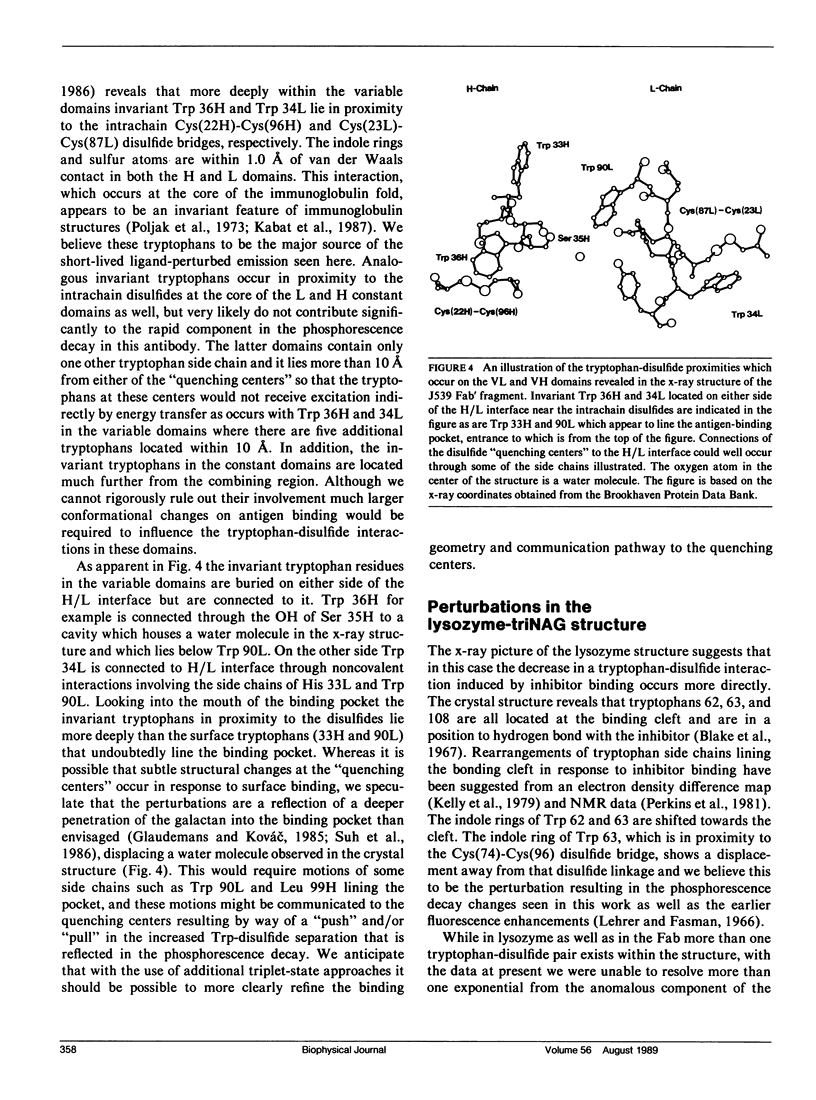
Phosphorescence spectroscopy on mouse myeloma IgA J539 in rigid solution at 77K revealed the type of anomalous short-lived component in the tryptophan decay originally observed with lysozyme (Churchich, J.E., 1966. Biochim. Biophys. Acta. 120:406-412) and seen in a large number of Bence Jones proteins (Longworth, J.W., C.L. McLaughlin, and A. Solomon. 1976. Biochemistry. 15:2953-2958). The decay time of the anomalous component that results from the interaction between tryptophan side chains and disulfide linkages in proteins was observed to significantly lengthen in J539 in response to binding of a galactan antigen. With hen egg-white lysozyme in which the type of fluorescence enhancement on ligand binding seen with J539 has also been observed, phosphorescence measurements revealed a similar lengthening of the decay time of the disulfide-induced anomalous component in the tryptophan decay. These perturbations are interpreted as ligand-induced changes to the tryptophan-disulfide proximities that have been shown to exist in these structures. Given the short-range nature of the disulfide perturbation (see following article) the observations suggest, in particular when combined with x-ray crystallographic data, that phosphorescence decay-time measurements of disulfide perturbations can serve as a sensitive spectroscopic indicator of subtle conformational changes in immunoglobulins and other tryptophan-disulfide containing proteins.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Blake C. C., Johnson L. N., Mair G. A., North A. C., Phillips D. C., Sarma V. R. Crystallographic studies of the activity of hen egg-white lysozyme. Proc R Soc Lond B Biol Sci. 1967 Apr 18;167(1009):378–388. doi: 10.1098/rspb.1967.0035. [DOI] [PubMed] [Google Scholar]
- Brewer J. M., Weber G. The effect of magnesium on some physical properties of yeast enolase. J Biol Chem. 1966 Jun 10;241(11):2550–2557. [PubMed] [Google Scholar]
- COWGILL R. W. Fluorescence and the structure of proteins. I. Effects of substituents on the fluorescence of indole and phenol compounds. Arch Biochem Biophys. 1963 Jan;100:36–44. doi: 10.1016/0003-9861(63)90031-6. [DOI] [PubMed] [Google Scholar]
- Churchich J. E. Tryptophan residues in native and reoxidized muramidase: luminescence properties. Biochim Biophys Acta. 1966 Jul 13;120(3):406–412. doi: 10.1016/0926-6585(66)90307-4. [DOI] [PubMed] [Google Scholar]
- Cowgill R. W. Fluorescence and the structure of proteins. 18. Spatial requirements for quenching by disulfide groups. Biochim Biophys Acta. 1970 Jun 23;207(3):556–559. doi: 10.1016/s0005-2795(70)80019-8. [DOI] [PubMed] [Google Scholar]
- Galley W. C., Stryer L. Triplet-singlet energy transfer in proteins. Biochemistry. 1969 May;8(5):1831–1838. doi: 10.1021/bi00833a008. [DOI] [PubMed] [Google Scholar]
- Galley W. C., Williams R. E., Goodfriend L. Unusual emission properties of the tryptophans at the surface of short ragweed allergen Ra5. Biochemistry. 1982 Jan 19;21(2):378–383. doi: 10.1021/bi00531a027. [DOI] [PubMed] [Google Scholar]
- Glaudemans C. P., Kovác P. Probing the combining site of monoclonal IgA J539 using deoxyfluoro- and other galactosides as ligands. Mol Immunol. 1985 Jun;22(6):651–653. doi: 10.1016/0161-5890(85)90094-x. [DOI] [PubMed] [Google Scholar]
- Glaudemans C. P., Kovác P., Rasmussen K. Mapping of subsites in the combining area of monoclonal anti-galactan immunoglobulin A J539. Biochemistry. 1984 Dec 18;23(26):6732–6736. doi: 10.1021/bi00321a069. [DOI] [PubMed] [Google Scholar]
- Hershberger M. V., Maki A. H. Heavy-atom effects associated with methylmercury(II) binding to rabbit glyceraldehyde-3-phosphate dehydrogenase. Biopolymers. 1980 Jul;19(7):1329–1344. doi: 10.1002/bip.1980.360190709. [DOI] [PubMed] [Google Scholar]
- Jolley M. E., Rudikoff S., Potter M., Glaudemans C. P. Spectral changes on binding of oligosaccharides to murine immunoglobulin A myeloma proteins. Biochemistry. 1973 Jul 31;12(16):3039–3044. doi: 10.1021/bi00740a015. [DOI] [PubMed] [Google Scholar]
- Kelly J. A., Sielecki A. R., Sykes B. D., James M. N., Phillips D. C. X-ray crystallography of the binding of the bacterial cell wall trisaccharide NAM-NAG-NAM to lysozyme. Nature. 1979 Dec 20;282(5741):875–878. doi: 10.1038/282875a0. [DOI] [PubMed] [Google Scholar]
- Khamis M. I., Casas-Finet J. R., Maki A. H. Stacking interactions of tryptophan residues and nucleotide bases in complexes formed between Escherichia coli single-stranded DNA binding protein and heavy atom-modified poly(uridylic) acid. A study by optically detected magnetic resonance spectroscopy. J Biol Chem. 1987 Feb 5;262(4):1725–1733. [PubMed] [Google Scholar]
- King L. A., Miller J. N. Factors affecting the luminescence of tryptophan at 77 K. Biochim Biophys Acta. 1976 Sep 28;446(1):206–213. doi: 10.1016/0005-2795(76)90111-2. [DOI] [PubMed] [Google Scholar]
- Kwiram A. L., Ross J. B. Optical detection of magnetic resonance in biologically important molecules. Annu Rev Biophys Bioeng. 1982;11:223–249. doi: 10.1146/annurev.bb.11.060182.001255. [DOI] [PubMed] [Google Scholar]
- Lehrer S. S., Fasman G. D. The fluorescence of lysozyme and lysozyme substrate complexes. Biochem Biophys Res Commun. 1966 Apr 19;23(2):133–138. doi: 10.1016/0006-291x(66)90517-1. [DOI] [PubMed] [Google Scholar]
- Longworth J. W., McLaughlin C. L., Solomon A. Luminescence studies on Bence-Jones proteins and light chains of immunoglobulins and their subunits. Biochemistry. 1976 Jul 13;15(14):2953–2958. doi: 10.1021/bi00659a003. [DOI] [PubMed] [Google Scholar]
- Miller J. N., King L. A. The low-temperature luminescence properties of bovine alpha-lactalbumin. Biochim Biophys Acta. 1975 Jun 26;393(2):435–445. doi: 10.1016/0005-2795(75)90072-0. [DOI] [PubMed] [Google Scholar]
- Monsigny M., Delmotte F., Hélène C. Ligands containing heavy atoms: perturbation of phosphorescence of a tryptophan residue in the binding site of wheat germ agglutinin. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1324–1328. doi: 10.1073/pnas.75.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins S. J., Johnson L. N., Phillips D. C., Dwek R. A. The binding of monosaccharide inhibitors to hen egg-white lysozyme by proton magnetic resonance at 270 MHz and analysis by ring-current calculations. Biochem J. 1981 Feb 1;193(2):553–572. doi: 10.1042/bj1930553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poljak R. J., Amzel L. M., Avey H. P., Chen B. L., Phizackerley R. P., Saul F. Three-dimensional structure of the Fab' fragment of a human immunoglobulin at 2,8-A resolution. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3305–3310. doi: 10.1073/pnas.70.12.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollet R., Edelhoch H. Changes in optical parameters of myeloma proteins with phosphorylcholine binding. J Biol Chem. 1974 Aug 25;249(16):5188–5194. [PubMed] [Google Scholar]
- Pollet R., Edelhoch H. The binding peoperties of anti-phosphorylcholine mouse myeloma proteins as measured by protein fluorescence. J Biol Chem. 1973 Aug 10;248(15):5443–5447. [PubMed] [Google Scholar]
- Purkey R. M., Galley W. C. Phosphorescence studies of environmental heterogeneity for tryptophyl residues in proteins. Biochemistry. 1970 Sep 1;9(18):3569–3575. doi: 10.1021/bi00820a010. [DOI] [PubMed] [Google Scholar]
- Satow Y., Cohen G. H., Padlan E. A., Davies D. R. Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A. J Mol Biol. 1986 Aug 20;190(4):593–604. doi: 10.1016/0022-2836(86)90245-7. [DOI] [PubMed] [Google Scholar]
- Saviotti M. L., Galley W. C. Room temperature phosphorescence and the dynamic aspects of protein structure. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4154–4158. doi: 10.1073/pnas.71.10.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambini G. B. Quenching of alkaline phosphatase phosphorescence by O2 and NO. Evidence for inflexible regions of protein structure. Biophys J. 1987 Jul;52(1):23–28. doi: 10.1016/S0006-3495(87)83184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh S. W., Bhat T. N., Navia M. A., Cohen G. H., Rao D. N., Rudikoff S., Davies D. R. The galactan-binding immunoglobulin Fab J539: an X-ray diffraction study at 2.6-A resolution. Proteins. 1986 Sep;1(1):74–80. doi: 10.1002/prot.340010112. [DOI] [PubMed] [Google Scholar]
- Vanderkooi J. M., Calhoun D. B., Englander S. W. On the prevalence of room-temperature protein phosphorescence. Science. 1987 May 1;236(4801):568–569. doi: 10.1126/science.3576185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang L. H., Maki A. H., Murphy J. B., Chase J. W. Triplet state sublevel kinetics of tryptophan 54 in the complex of Escherichia coli single-stranded DNA binding protein with single-stranded poly(deoxythymidylic) acid. Biophys J. 1987 Nov;52(5):867–872. doi: 10.1016/S0006-3495(87)83280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuclich J. A., Maki A. H. Protein triplet states. Top Curr Chem. 1975;(54):115–163. doi: 10.1007/BFb0046182. [DOI] [PubMed] [Google Scholar]


