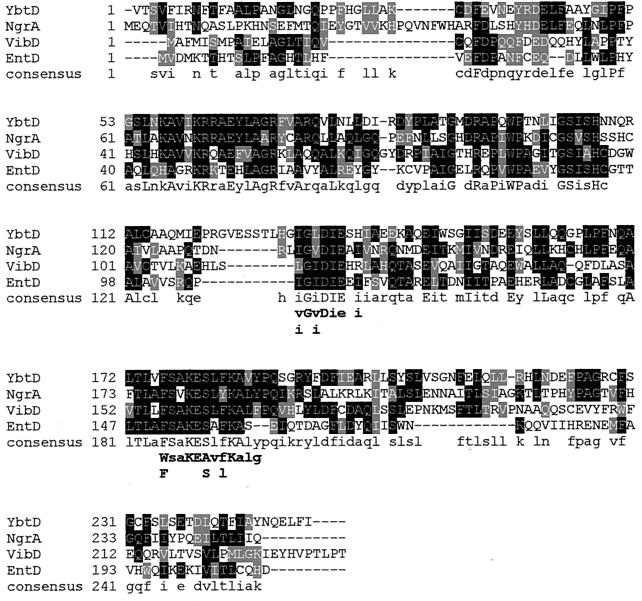
FIG. 3.
Amino acid sequence alignment of YbtD from Y. pestis, NgrA from P. luminescens, VibD from V. cholerae, and EntD from E. coli. Residues with identity to YbtD are in white with a solid black background. Conservative and semiconservative amino acid substitutions are shaded. The consensus line shows identical residues in all four proteins (uppercase letters) and identical residues in two or more proteins (lowercase letters). The identity and similarity of YbtD to each of these proteins are 34 and 65.3% (NgrA), 31.3 and 60.2% (VibD), and 27.2 and 58.5% (EntD). The residues below the consensus line indicate conserved (lowercase) and highly conserved (uppercase) amino acids within the proposed P-pant transferase domain derived from comparison of 22 P-pant transferases (46).