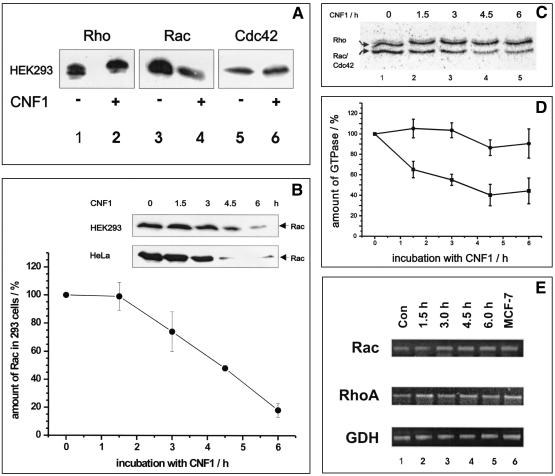
FIG. 1.
(A) Detection of Rho proteins from CNF1-treated cells by Western blotting. HEK293 cells were treated with 500 ng of CNF1 per ml overnight and lysed in boiling SDS buffer. The crude lysates were separated by SDS-PAGE and transferred onto a nitrocellulose membrane. Rho GTPases were detected with specific antibodies. Typical results of three different experiments are shown. (B) Time course of Rac degradation in CNF1-treated cells. HEK293 and HeLa cells were incubated in the presence or absence of 500 ng of CNF1 per ml for different times. The cells were lysed in boiling SDS buffer, and the crude lysates were separated by SDS-PAGE. Proteins were transferred onto a nitrocellulose membrane and probed for Rac with a specific Rac antibody. Bands obtained in three independent experiments (HEK293 cells) were quantified by using the ImageQuant 5.2 software. The amount of Rac detected in lysates of HEK293 cells without CNF1 treatment (zero time) was defined as 100%. The means ± standard deviations of three independent experiments are shown. (C) Detection of Rho proteins from CNF1-treated cells by glucosylation with toxin B. HEK293 cells were treated with 500 ng of CNF1 per ml and lysed in lysis buffer. Lysates were treated with C. difficile toxin B in the presence of labeled UDP-[14C]glucose. Proteins were separated by SDS-PAGE. Gels were stained and dried, and glucosylated proteins were detected by phosphorimaging. Note that the upper band shifted to a higher molecular mass, indicating that there was deamidation of RhoA by CNF1. The lower band represents mostly Rac but also a small amount of Cdc42. Typical results of four different experiments are shown. (D) Quantification of Rho proteins. For quantification of the labeled RhoA and Rac-Cdc42 bands, the ImageQuant software (Amersham Pharmacia Biotech) was used. The means ± standard deviations of four different experiments are shown. (E) Analysis of Rho mRNA expression. In order to analyze expression of the diverse Rho species at the mRNA level, total RNA was isolated from untreated (Con; lane 1) and CNF-treated (lanes 2 to 5) cells and cDNA was synthesized. Two microliters of the first-strand reaction mixture was subjected to a standard PCR. GDH was used as the internal standard. After PCR the products were separated on 1.5% agarose gels and visualized by ethidium bromide staining. Lysate of MCF-7 (a human breast carcinoma cell line) was used as an internal control for the assay conditions (lane 6).