Abstract
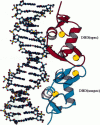
Molecular dynamics simulations have been performed on the glucocorticoid receptor DNA binding domain (GR DBD) in aqueous solution as a dimer in complex with DNA and as a free monomer. In the simulated complex, we find a slightly increased bending of the DNA helix axis compared with the crystal structure in the spacer region of DNA between the two half-sites that are recognized by GR DBD. The bend is mainly caused by an increased number of interactions between DNA and the N-terminal extended region of the sequence specifically bound monomer. The recognition helices of GR DBD are pulled further into the DNA major groove leading to a weakening of the intrahelical hydrogen bonds in the middle of the helices. Many ordered water molecules with long residence times are found at the intermolecular interfaces of the complex. The hydrogen-bonding networks (including water bridges) on either side of the DNA major groove involve residues that are highly conserved within the family of nuclear receptors. Very similar hydrogen-bonding networks are found in the estrogen receptor (ER) DBD in complex with DNA, which suggests that this is a common feature for proper positioning of the recognition helix in ER DBD and GR DBD.
Full text
PDF
























Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Bartenev V. N., Golovamov EuI, Kapitonova K. A., Mokulskii M. A., Volkova L. I., Skuratovskii IYa Structure of the B DNA cationic shell as revealed by an X-ray diffraction study of CsDNA. Sequence-specific cationic stabilization of B form DNA. J Mol Biol. 1983 Sep 5;169(1):217–234. doi: 10.1016/s0022-2836(83)80181-8. [DOI] [PubMed] [Google Scholar]
- Baumann H., Paulsen K., Kovács H., Berglund H., Wright A. P., Gustafsson J. A., Härd T. Refined solution structure of the glucocorticoid receptor DNA-binding domain. Biochemistry. 1993 Dec 14;32(49):13463–13471. doi: 10.1021/bi00212a011. [DOI] [PubMed] [Google Scholar]
- Beato M. Gene regulation by steroid hormones. Cell. 1989 Feb 10;56(3):335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- Berglund H., Kovács H., Dahlman-Wright K., Gustafsson J. A., Härd T. Backbone dynamics of the glucocorticoid receptor DNA-binding domain. Biochemistry. 1992 Dec 8;31(48):12001–12011. doi: 10.1021/bi00163a007. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Brunne R. M., Liepinsh E., Otting G., Wüthrich K., van Gunsteren W. F. Hydration of proteins. A comparison of experimental residence times of water molecules solvating the bovine pancreatic trypsin inhibitor with theoretical model calculations. J Mol Biol. 1993 Jun 20;231(4):1040–1048. doi: 10.1006/jmbi.1993.1350. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Karplus M. Polar hydrogen positions in proteins: empirical energy placement and neutron diffraction comparison. Proteins. 1988;4(2):148–156. doi: 10.1002/prot.340040208. [DOI] [PubMed] [Google Scholar]
- Clark K. L., Halay E. D., Lai E., Burley S. K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993 Jul 29;364(6436):412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Bax A., Omichinski J. G., Gronenborn A. M. Localization of bound water in the solution structure of a complex of the erythroid transcription factor GATA-1 with DNA. Structure. 1994 Feb 15;2(2):89–94. doi: 10.1016/s0969-2126(00)00011-3. [DOI] [PubMed] [Google Scholar]
- Dahlman-Wright K., Wright A., Gustafsson J. A., Carlstedt-Duke J. Interaction of the glucocorticoid receptor DNA-binding domain with DNA as a dimer is mediated by a short segment of five amino acids. J Biol Chem. 1991 Feb 15;266(5):3107–3112. [PubMed] [Google Scholar]
- Danielsen M., Hinck L., Ringold G. M. Two amino acids within the knuckle of the first zinc finger specify DNA response element activation by the glucocorticoid receptor. Cell. 1989 Jun 30;57(7):1131–1138. doi: 10.1016/0092-8674(89)90050-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Eriksson M. A., Berglund H., Härd T., Nilsson L. A comparison of 15N NMR relaxation measurements with a molecular dynamics simulation: backbone dynamics of the glucocorticoid receptor DNA-binding domain. Proteins. 1993 Dec;17(4):375–390. doi: 10.1002/prot.340170406. [DOI] [PubMed] [Google Scholar]
- Eriksson M. A., Laaksonen A. A molecular dynamics study of conformational changes and hydration of left-handed d(CGCGCGCGCGCG)2 in a nonsalt solution. Biopolymers. 1992 Aug;32(8):1035–1059. doi: 10.1002/bip.360320814. [DOI] [PubMed] [Google Scholar]
- Härd T., Kellenbach E., Boelens R., Kaptein R., Dahlman K., Carlstedt-Duke J., Freedman L. P., Maler B. A., Hyde E. I., Gustafsson J. A. 1H NMR studies of the glucocorticoid receptor DNA-binding domain: sequential assignments and identification of secondary structure elements. Biochemistry. 1990 Sep 25;29(38):9015–9023. doi: 10.1021/bi00490a019. [DOI] [PubMed] [Google Scholar]
- Härd T., Kellenbach E., Boelens R., Maler B. A., Dahlman K., Freedman L. P., Carlstedt-Duke J., Yamamoto K. R., Gustafsson J. A., Kaptein R. Solution structure of the glucocorticoid receptor DNA-binding domain. Science. 1990 Jul 13;249(4965):157–160. doi: 10.1126/science.2115209. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Fratini A. V., Drew H. R., Dickerson R. E. Ordered water structure around a B-DNA dodecamer. A quantitative study. J Mol Biol. 1983 Jan 5;163(1):129–146. doi: 10.1016/0022-2836(83)90033-5. [DOI] [PubMed] [Google Scholar]
- Laudet V., Hänni C., Coll J., Catzeflis F., Stéhelin D. Evolution of the nuclear receptor gene superfamily. EMBO J. 1992 Mar;11(3):1003–1013. doi: 10.1002/j.1460-2075.1992.tb05139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J Biomol Struct Dyn. 1988 Aug;6(1):63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Levitt M., Park B. H. Water: now you see it, now you don't. Structure. 1993 Dec 15;1(4):223–226. doi: 10.1016/0969-2126(93)90011-5. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991 Aug 8;352(6335):497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Mader S., Kumar V., de Verneuil H., Chambon P. Three amino acids of the oestrogen receptor are essential to its ability to distinguish an oestrogen from a glucocorticoid-responsive element. Nature. 1989 Mar 16;338(6212):271–274. doi: 10.1038/338271a0. [DOI] [PubMed] [Google Scholar]
- Misra V. K., Sharp K. A., Friedman R. A., Honig B. Salt effects on ligand-DNA binding. Minor groove binding antibiotics. J Mol Biol. 1994 Apr 29;238(2):245–263. doi: 10.1006/jmbi.1994.1285. [DOI] [PubMed] [Google Scholar]
- Otting G., Liepinsh E., Wüthrich K. Protein hydration in aqueous solution. Science. 1991 Nov 15;254(5034):974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Ravishanker G., Swaminathan S., Beveridge D. L., Lavery R., Sklenar H. Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCGAATTCGCG) double helix: "curves", dials and windows. J Biomol Struct Dyn. 1989 Feb;6(4):669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- Schena M., Freedman L. P., Yamamoto K. R. Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA binding and transcriptional enhancement activities. Genes Dev. 1989 Oct;3(10):1590–1601. doi: 10.1101/gad.3.10.1590. [DOI] [PubMed] [Google Scholar]
- Schwabe J. W., Chapman L., Finch J. T., Rhodes D., Neuhaus D. DNA recognition by the oestrogen receptor: from solution to the crystal. Structure. 1993 Nov 15;1(3):187–204. doi: 10.1016/0969-2126(93)90020-h. [DOI] [PubMed] [Google Scholar]
- Schwabe J. W., Chapman L., Finch J. T., Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993 Nov 5;75(3):567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- Schwabe J. W., Neuhaus D., Rhodes D. Solution structure of the DNA-binding domain of the oestrogen receptor. Nature. 1990 Nov 29;348(6300):458–461. doi: 10.1038/348458a0. [DOI] [PubMed] [Google Scholar]
- Seibel G. L., Singh U. C., Kollman P. A. A molecular dynamics simulation of double-helical B-DNA including counterions and water. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6537–6540. doi: 10.1073/pnas.82.19.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Y., Carlstedt-Duke J., Weigel N. L., Dahlman K., Gustafsson J. A., Tsai M. J., O'Malley B. W. Molecular interactions of steroid hormone receptor with its enhancer element: evidence for receptor dimer formation. Cell. 1988 Oct 21;55(2):361–369. doi: 10.1016/0092-8674(88)90059-1. [DOI] [PubMed] [Google Scholar]
- Umesono K., Evans R. M. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989 Jun 30;57(7):1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- Van Gunsteren W. F., Berendsen H. J., Geurtsen R. G., Zwinderman H. R. A molecular dynamics computer simulation of an eight-base-pair DNA fragment in aqueous solution: comparison with experimental two-dimensional NMR data. Ann N Y Acad Sci. 1986;482:287–303. doi: 10.1111/j.1749-6632.1986.tb20962.x. [DOI] [PubMed] [Google Scholar]
- Zilliacus J., Dahlman-Wright K., Wright A., Gustafsson J. A., Carlstedt-Duke J. DNA binding specificity of mutant glucocorticoid receptor DNA-binding domains. J Biol Chem. 1991 Feb 15;266(5):3101–3106. [PubMed] [Google Scholar]
- Zilliacus J., Wright A. P., Norinder U., Gustafsson J. A., Carlstedt-Duke J. Determinants for DNA-binding site recognition by the glucocorticoid receptor. J Biol Chem. 1992 Dec 15;267(35):24941–24947. [PubMed] [Google Scholar]
- de Vlieg J., Berendsen H. J., van Gunsteren W. F. An NMR-based molecular dynamics simulation of the interaction of the lac repressor headpiece and its operator in aqueous solution. Proteins. 1989;6(2):104–127. doi: 10.1002/prot.340060203. [DOI] [PubMed] [Google Scholar]