Figure 1.
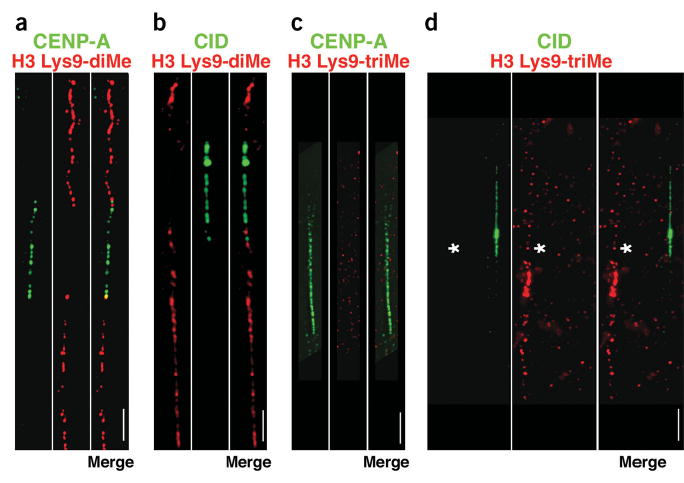
H3 is not di- or trimethylated at Lys9 in CEN chromatin. (a–d) Extended chromatin fibers from human (a,c) or D. melanogaster (b,d) interphase cells were stained with antibodies to CENP-A or CID (green), and H3 Lys9-diMe or H3 Lys9-triMe (red), detected with FITC- and Cy3-conjugated secondary antibodies. Merged images are to the right of the single-color images. Spaces between CENP-A/CID blocks denote H3-containing nucleosome blocks15, and did not stain for H3 Lys9-diMe or H3 Lys9-triMe. H3 Lys9-diMe, typically found in heterochromatin, was present on one side (67%, n = 30) or both sides (33%; n = 30) of the human CENP-A region (a). Lys9 was not dimethylated on H3 within the CID domain on chromatin fibers from D. melanogaster third-instar larval neuroblasts and S2 cells, and always flanked the CID region on both sides (b). In contrast, H3 Lys9-triMe was not present in the regions flanking CENP-A/CID (c,d). To demonstrate that staining for H3 Lys9-triMe was present in noncentromeric regions in these preparations, two different fibers are shown in d, one that is CID-negative but H3 Lys9-triMe-positive (denoted by asterisk), and one that is CID-positive and H3 Lys9-triMe-negative. Quantitative evaluations of the overlaps are presented in Figure 2. Scale bars, 15 μm in a,c; 5 μm in b,d.
