Abstract
The major structural determinant of the preference to bind to CpG binding sites on DNA exhibited by the natural quinoxaline bis-intercalators echinomycin and triostin A, or the quinoline echinomycin derivative, 2QN, is the 2-amino group of guanine (G). However, relocation of this group by means of introduction into the DNA molecule of the 2-aminoadenine (=2,6-diaminopurine, D) base in place of adenine (A) has been shown to lead to a drastic redistribution of binding sites, together with ultratight binding of 2QN to the sequence DTDT. Also, the demethylated triostin analogs, TANDEM and CysMeTANDEM, which bind with high affinity to TpA steps in natural DNA, bind much less tightly to CpI steps, despite the fact that both adenosine and the hypoxanthine-containing nucleoside, inosine (I), provide the same hydrogen bonding possibilities in the minor groove. To study both the increased binding affinity of 2QN for DTDT relative to GCGC sites and the remarkable loss of binding energy between CysMeTANDEM and ICIC compared with ATAT, a series of thermodynamic integration free energy simulations involving conversions between DNA base pairs have been performed. Our results demonstrate that the electrostatic component of the stacking interactions between the heteroaromatic rings of these compounds and the bases that make up the intercalation sites plays a very important role in the modulation of their binding affinities.
INTRODUCTION
The ability of a given stretch of DNA to act as a molecular recognition target appears, at first sight, to be limited to functional group discrimination along the major and minor grooves that lie between the phosphodiester linkages of both strands (1). While this is true for many sequence-specific DNA-binding proteins and low-molecular-weight ligands [including natural and synthetic information-reading lexitropsins (2), as well as many clinically used antitumor drugs (3)], it is also perceived nowadays that additional potential for specific recognition is provided by sequence-dependent DNA microheterogeneity (4), which is mostly due to differences in stacking interactions between adjacent base pairs. These differences translate, for example, into differential propensities to bend (5) and preferential intercalation of planar heteroaromatic ring systems at particular base pair steps (6,7).
Interest in bifunctional intercalators stems not only from the possibility of enhancing their binding affinity over that of the corresponding monomers but also from the greater opportunities for imposing selective binding to defined sequences afforded by the bracketing of two (or more) base pairs between the intercalation sites. Thus, for a binding site covering 4 bp the number of distinguishable sequences is 136 versus only 10 unique dinucleotide steps at which monointercalation can take place.
Nature provides some remarkable examples of bifunctional intercalators in the family of quinoxaline antibiotics represented by echinomycin [a.k.a. quinomycin A] and its biosynthetic precursor, triostin A, that are primarily produced, respectively, by Streptomyces echinatus and Streptomyces triostinicus, and for which a binding site of 4 bp was early demonstrated and then repeatedly confirmed (8). Interestingly, these natural bis-intercalators exhibit a definite preference for binding to a 5′-CpG-3′ core flanked by an A:T pair on either side, as opposed to other natural crescent-shaped non-intercalating ligands (e.g. netropsin and distamycin), which show a strong preference for binding to the minor groove of A,T-rich DNA regions (9).
Traditional work in structure-affinity relationships (SAR) for quinoxaline antibiotics has dealt with the effects that introduction of new substituents or removal of existing ones have on the binding properties of the parent compounds. Changes in binding affinity and specificity have been observed for derivatives modified on either the intercalating bicycle (8,10) or the octadepsipeptide ring (8). On the other hand, structural studies using both X-ray (11,12) and NMR (13–16) techniques have shed light into the molecular basis of the interaction between several of these ligands and short oligonucleotides containing some of their referred binding sites. Thus, these staple-shaped antibiotics have been shown to bind to DNA making use of both nearest-neighbor-exclusion-compliant (1) bifunctional intercalation of their heteroaromatic chromophores and important minor-groove contacts between the two sandwiched DNA base pairs and the cyclic depsipeptide linker, the most crucial being a number of hydrogen bonds formed between the NH and CO groups of alanine and the N3 and NH2 groups of the guanines making up the central CpG step.
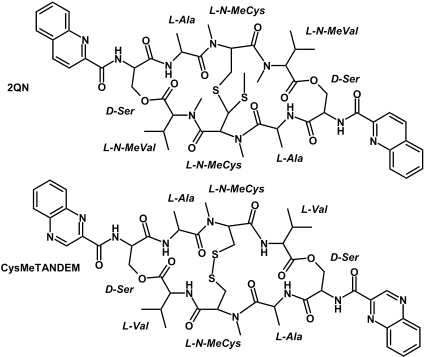
The analogs that have been most intensively studied are 2QN, the bis-quinoline derivative of echinomycin (8,10), and des-N-tetramethyl triostin A (TANDEM) (17,18) and [N-MeCys3,N-MeCys7] TANDEM (CysMeTANDEM) (19–22), which lack, respectively, either all or half of the N-methyl groups of cysteines and valines present in the cyclic depsipeptide of triostin A (Figure 1). Although no great differences from echinomycin are found for 2QN (10), total or partial N-demethylation brings about a rather drastic change in binding specificity as both TANDEM and CysMeTANDEM bind to TpA steps in place of the CpG steps recognized by both triostin A and echinomycin in standard DNA (8,18,20). This has been rationalized on the basis of a loss of hydrogen bonding possibilities with the 2-amino group of guanines due to the formation of two intramolecular hydrogen bonds between the CO of alanines and the NH of valines in TANDEM and CysMeTANDEM.
Figure 1.
Chemical structures and amino acid composition of DNA bis-intercalators 2QN (top) and CysMeTANDEM (bottom).
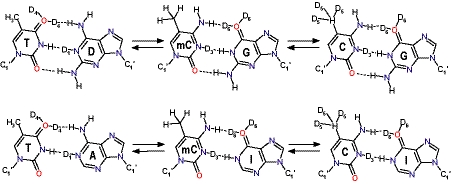
The availability of DNA molecules containing modified purine bases such as hypoxanthine (resulting in the nucleoside inosine, I) and 2,6-diaminopurine (DAP, D) (Figure 2) allowed an interesting extension of the SAR studies (23) and provided new insight into the molecular determinants of binding selectivity (24). As expected, due to the reported ascendancy of the exocyclic 2-amino group in the binding of these compounds, it was found that replacement of all Gs by Is resulted in triostin A (25), echinomycin (26) and 2QN (10) completely failing to bind to these modified nucleotides. Strikingly, however, replacing all As with Ds led to an unexpected redistribution of binding sites for echinomycin (23) and 2QN (10) to other pyrimidine–purine combinations, such as TpD and CpD, in preference to the usual CpG step despite the fact that, from a minor-groove viewpoint, CpG and TpD steps share the same hydrogen bonding capabilities (Figure 2). Moreover, remarkably tight binding was observed between 2QN and a particular DTDT sequence in the plasmid studied (10). More recently, measurements of contour lengths provided by atomic force microscopy images of linear and circular DNA have been used to confirm the much higher affinity of echinomycin for D-substituted and (I+D)-substituted DNA fragments over natural DNA (27).
Figure 2.
Schematic representation of the DNA base pairs and dummy atoms used in the MD-TI simulations. mC stands for 5-methyl-cytosine. Dummy atoms labels correspond with those of atoms to which they are attached.
Equally intriguing are the findings that the G→I replacement in the DNA molecule has no influence on the interaction of TANDEM with TpA steps (i.e. TANDEM does not bind to CpI steps) (25) and, similarly, that binding of CysMeTANDEM to CpI is much weaker than binding to the high-affinity TpA sites (16) despite the fact that the minor grooves of CpI and TpA steps provide the same hydrogen bonding possibilities (Figure 2). Furthermore, the affinity of CysMeTANDEM (21,22) and TANDEM (8,18) for binding TpA-containing sequences is clearly affected by the flanking bases, with the tetranucleotides ATAT and TTAA being the best and poorest binding sites, respectively.
These observations raise some questions as to the origin of the binding specificity, which appears to depend on some additional factors beyond hydrogen bonding interactions between the depsipeptide and the minor groove. Since the bis-intercalative interaction of echinomycin has been shown to be entropically driven (28), suggesting that the process is predominantly stabilized by hydrophobic interactions, it has been hypothesized that the binding selectivity of these compounds might originate from differences in steric and/or hydrophobic interactions with a minor groove of suitable dimensions rather than with specific hydrogen bond formation (25). Nonetheless, two independent lines of evidence appear to suggest that aromatic stacking interactions can be particularly relevant in determining the sequence specificity for this group of bis-intercalating ligands. On the one hand, data provided by low-temperature phosphorescence and triplet-state magnetic resonance spectroscopy revealed a correlation between the stabilization of the complexes of either echinomycin (29) or 2QN (30) with several double-stranded DNA molecules and the extent of stacking interactions of their chromophores. On the other hand, energy analyses of computer-generated molecular models of a series of 1:1 complexes between echinomycin and DNA oligonucleotides containing standard and modified nucleobases revealed the distinct stacking properties of D:T and G:C base pairs (31).
To further test the hypothesis that stacking interactions can play a decisive role in modulating the preferential binding of these bis-intercalating ligands, we have used free energy simulations to study the origins of both the remarkably increased affinity of 2QN for DTDT relative to GCGC sites and the notable loss of binding affinity between CysMeTANDEM and ICIC compared with ATAT. This strategy was successfully used in the computational study of netropsin binding to the minor grooves of poly[d(IC)]·poly[d(IC)] and poly[d(GC)]·poly[d(GC)] (32) although in this early case the ‘mutation’ only affected the purine base. In the present simulations, mutations involve the whole base pairs, and calculations are performed in such a way that the contributions of the base pairs on either side of the intercalated chromophores can be separately assessed.
MATERIALS AND METHODS
Force field and charges
Electrostatic potential-derived (ESP) charges for the non-standard residues making up 2QN and CysMeTANDEM (L-N-MeCys, L-N-MeVal, D-Ser, N-methylquinoline-2-carboxamide and N-methylquinoxaline-2-carboxamide) as well as for the N9-methylated derivatives of 2,6-diaminopurine and hypoxanthine, were obtained with the RESP methodology (33) and the wave functions determined at the HF/6-31G* level using Gaussian-98 program (34). The suitability of this procedure is supported by the excellent agreement found between the dipole moment calculated from ESP charges for N-methyl-quinoxaline-2-carboxamide (μ = 4.14 D) and its experimentally determined value (μ = 4.15 ± 0.03 D) (31). Point charges for amide atoms in the depsipeptide part of the ligands were restrained to the values these atoms have in the AMBER force field (35). Additional bonded and non-bonded parameters (Supplementary Data) were derived, by analogy or through interpolation, from those already present in the AMBER database (parm99.dat).
Construction of the starting structures
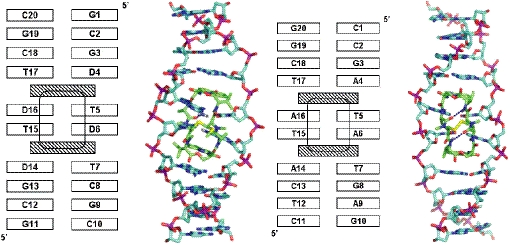
Models for the free oligonucleotides were built using optimized parameters for B-DNA (36). A previously reported 2QN:d(GACGTC)2 model (10) was used to build the complex of 2QN with the decanucleotide d(GCGDTDTCGC)2, which corresponds to a strongly protected site identified at position 76 in the DAP-containing 160 bp TyrT DNA fragment used in footprinting experiments (10) (base pairs sandwiched by the drug chromophores are shown in bold). The template for the CysMeTANDEM:d(CTCATATCAG)2 complex (also a strongly protected site determined experimentally) (20) was the NMR solution structure (14) [PDB entry 2DA8] of the complex formed between this drug and the hexanucleotide d(GATATC)2 that yielded the lowest potential energy and the best intermolecular hydrogen-bonding scheme, as reported previously (31). Appropriate modifications of base composition were introduced by using standard geometries (36) and replacing the respective purine and pyrimidine bases where necessary. In both cases, the central tetranucleotide where the most intimate interactions with the drug take place is embedded in a DNA decamer so as to better simulate the experimental conditions (Figure 3). In addition, terminal G:C base pairs for both 5′ and 3′ ends were used to avoid any possible end-effects.
Figure 3.
Schematic and stick representation of the initial complexes studied for 2QN (left) and CysMeTANDEM (right) showing base composition and numbering. The intramolecular hydrogen bonds present in CysMeTANDEM are displayed as dotted lines.
Molecular dynamics simulations
Each molecular system was neutralized by the addition of 18 sodium ions (37), placed along the phosphate bisectors, and immersed in a rectangular box of 4572 TIP3P water molecules (38). Periodic boundary conditions were applied and electrostatic interactions were treated using the smooth particle mesh Ewald method (39) with a grid spacing of ∼1 Å. The cutoff distance for the non-bonded interactions was 9 Å. The SHAKE algorithm (40) was applied to all bonds and an integration step of 1.0 fs was used throughout. Solvent molecules and counterions were relaxed by energy minimization and allowed to redistribute around the positionally restrained solute (25 kcal mol−1 Å−2) during 50 ps of molecular dynamics (MD) at constant temperature (300 K) and pressure (1 atm). These initial harmonic restraints were gradually reduced until their complete removal in a series of progressive energy minimizations. The resulting systems were then heated from 100 to 300 K during 10 ps and subsequently allowed to equilibrate for 1 ns of unrestrained MD. System coordinates were saved every 2.0 ps for further analysis.
Calculation of binding free energy differences
Thermodynamic integration (TI) calculations (41), as implemented in the Gibbs module of AMBER 6 (http://amber.scripps.edu/doc6/amber6.pdf), were used to investigate the free energy differences for 2QN binding to DTDT and GCGC sequences and for CysMeTANDEM binding to ATAT and ICIC steps. In this method, the system is ‘mutated’ from one state to another with the aid of dummy atoms (D1, D3, D4, D5 and D6 in Figure 2) by changing the interaction parameters that define the Hamiltonian H as a function of a coupling parameter, λ. The free energy difference, ΔG, between state A, defined by H(λ = 0) = HA, and state B, defined by H(λ = 1) = HB, is calculated as follows:
| 1 |
where 〈〉λ denotes ensemble averaging at a given value of λ, and the Hamiltonian of the system at any intermediate state, H[λ], is defined as (1 − λ)HA + λHB.
The integrand in Equation 1 is evaluated numerically from MD simulations at specified values of λ and the total integral is approximated from these data using trapezoidal numerical integration. Simulation conditions were identical to those used in the equilibration MD runs. We initially used a total of 41 equally spaced integration points (‘windows’) for both the ‘forward’ (λ = 1 to λ = 0) and ‘backward’ (λ = 0 to λ = 1) directions, each consisting of 5 ps of equilibration and 5 ps of data collection. To check for protocol dependencies, these times were then doubled to 10 ps each in a second set of simulations. Consequently, a total of four perturbations were done for each molecular system, and the averages were used to assess the convergence of the results and get a crude estimate of the net error.
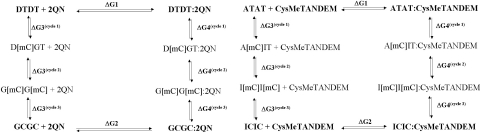
The whole thermodynamic cycle for each decanucleotide or drug:decanucleotide complex was initially divided into two parts in order to examine separately the contributions to the total free energy change of flanking and central base pairs. Nonetheless, test simulations for the DTDT-containing sequence showed that mutation of thymine to cytosine (and vice versa) required consideration of 5-methyl-cytosine (mC) as an intermediate state (Figure 2) to reduce convergence problems (42) associated with the bonded contributions (ΔGpmf) due to the growth/disappearance of the methyl group and the simultaneous interconversion of the vicinal keto group to an amino group. Thus, each thermodynamic cycle was split into three independent perturbations (Figure 4). First, the central T:D or T:A base pairs sandwiched by the drug were converted to either mC:G or mC:I, respectively; then, the T:D or T:A base pairs flanking the bis-intercalation site were mutated to either mC:G or mC:I; and, finally, the methyl group of all mCs was mutated to hydrogen.
Figure 4.
Thermodynamic cycles used for the computation of the free energy differences.
After 1 ns of MD equilibration for both the DTDT free decamer and the 2QN:DTDT complex, the TI simulation DTDT→D[mC]GT was performed. At the end of the perturbation, the final state was equilibrated for 1 ns before undertaking both the backward simulation, i.e. D[mC]GT→DTDT, and the next forward perturbation leading from D[mC]GT to G[mC]G[mC]. At the end of this second cycle, the G[mC]G[mC]-containing decanucleotide was further equilibrated for 1 ns of MD before the resulting structure was used to run both the backward perturbation (G[mC]G[mC]→D[mC]GT) and the next and final forward simulation (third cycle) leading from G[mC]G[mC] to GCGC. An identical protocol was used in the MD-TI simulations performed for CysMeTANDEM.
The relative free energy difference for the binding of either 2QN to DTDT and GCGC or CysMeTANDEM to ATAT and ICIC, respectively, was calculated as
| 2 |
Analysis of the molecular dynamics trajectories and electrostatic energy calculations
Three-dimensional structures and trajectories were visually inspected using computer graphics. Root-mean-square deviations (rmsd) from the initial structures and interatomic distances were monitored using the CARNAL module in AMBER. Finite-difference solutions to the linearized Poisson–Boltzmann equation, as implemented in the DelPhi program (version 2.5), were used to compute the electrostatic components of the ligand–DNA binding free energy, including the desolvation cost, using snapshots from the MD simulations and following the procedure described in detail elsewhere (43,44). AMBER charges and radii were employed, and the solute boundaries were defined by calculating the solvent-accessible surfaces with a spherical probe with a radius of 1.4 Å. A minimum separation of 10 Å was left between any solute atom and the borders of the box. The potentials at the grid points delimiting the box were calculated analytically by treating each charge atom as a Debye–Hückel sphere. The interior of both the DNA and the ligand was considered a low-dielectric medium (ɛ = 2), whereas the surrounding solvent was treated as a high-dielectric medium (ɛ = 80) with an ionic strength of 0.145 M.
Targeted molecular dynamics simulations
A targeted molecular dynamics (tMD) approach was used to compare the relative energetic costs involved in creation of the two intercalation sites in the four decanucleotides studied. The methodology was essentially the same as previously described for creation of a ligand-binding cavity in the HIV-1 reverse transcriptase enzyme (45) and made use of the standard implementation recently incorporated into AMBER 8.0 (http://amber.scripps.edu/doc8/amber8.pdf). A restraint was defined in terms of a mass-weighted root-mean-square (rms) superposition to the final reference structure (target) and applied in the force field as an extra energy term of the following form:
| 3 |
where kr is the force constant, N is the number of atoms, and trmsd is the target rms deviation, which was set to zero.
A force constant of 1.0 kcal mol−1 Å−2 over 0.5 ns or 5 ns proved sufficient to find a low-energy path leading from the free DNA decamer to the target structure (the same decamer as found in its complex with the bis-intercalating ligand) using only the DNA heavy atoms in the rms definition.
RESULTS AND DISCUSSION
Differences in binding affinities of 2QN and CysMeTANDEM
Each of the two DNA decamers containing a central high-affinity target site for either 2QN or CysMeTANDEM was converted to an alternative decamer containing a central low-affinity site both in the free state and in complex with the respective bis-intercalating ligand. It can be seen (Tables 1 and 2) that the largest error estimate calculated by averaging the results determined from four MD-TI simulations is typically on the order of 0.3 kcal mol−1 and never greater than 0.9 kcal mol−1, which lends credence to the convergence of the results. In both sets of perturbations, the largest free energy changes (cycle 3) corresponded to the shrinkage of the methyl group to a hydrogen (or the reverse growth of a hydrogen to a methyl) even though the computed differences were very similar for the free and complexed DNA. On the other hand, the energy decomposition showed that the largest difference in all cases originated from the electrostatic term (ΔGele).
Table 1.
Averaged differences in free energy components (kcal mol−1) calculated for the binding of 2QN to the DTDT- and GCGC-containing decanucleotides according to the thermodynamic cycles depicted in Figure 4
| (A) | ΔG(4)cycle1 | ΔG(3)cycle1 |
| 2QN:DTDT→2QN:D[mC]GT | DTDT→D[mC]GT | |
| ΔGtot | 82.8 ± 0.3 | 81.6 ± 0.3 |
| ΔGele | 77.5 ± 0.2 | 76.9 ± 0.1 |
| ΔGvdW | 0.5 ± 0.0 | 0.5 ± 0.1 |
| ΔGpmf | 4.8 ± 0.1 | 4.5 ± 0.2 |
| (B) | ΔG(4)cycle2 | ΔG(3)cycle2 |
| 2QN:D[mC]GT→2QN:G[mC]G[mC] | D[mC]GT→G[mC]G[mC] | |
| ΔGtot | 79.3 ± 0.1 | 77.7 ± 0.1 |
| ΔGele | 74.8 ± 0.2 | 72.7 ± 0.1 |
| ΔGvdW | 0.1 ± 0.1 | 0.1 ± 0.1 |
| ΔGpmf | 4.7 ± 0.1 | 5.0 ± 0.1 |
| (C) | ΔG(4)cycle3 | ΔG(3)cycle3 |
| 2QN:G[mC]G[mC]→2QN:GCGC | G[mC]G[mC]→GCGC | |
| ΔGtot | 353.8 ± 0.1 | 354.8 ± 0.1 |
| ΔGele | 346.5 ± 0.1 | 346.0 ± 0.1 |
| ΔGvdW | 0.7 ± 0.1 | 2.1 ± 0.1 |
| ΔGpmf | 6.8 ± 0.1 | 7.0 ± 0.1 |
Table 2.
Averaged differences in free energy components (kcal mol−1) calculated for the binding of CysMeTANDEM to the ATAT- and ICIC-containing decanucleotides according to the thermodynamic cycles depicted in Figure 4
| (A) | ΔG(4)cycle1 | ΔG(3)cycle1 |
| CysMeTANDEM:ATAT→CysMeTANDEM:A[mC]IT | ATAT→A[mC]IT | |
| ΔGtot | 50.5 ± 0.8 | 48.7 ± 0.8 |
| ΔGele | 46.2 ± 0.8 | 45.6 ± 0.4 |
| ΔGvdW | 0.3 ± 0.1 | 0.8 ± 0.1 |
| ΔGpmf | 4.0 ± 0.1 | 2.4 ± 0.5 |
| (B) | ΔG(4)cycle2 | ΔG(3)cycle2 |
| CysMeTANDEM:A[mC]IT→CysMeTANDEM:I[mC]I[mC] | A[mC]IT→I[mC]I[mC] | |
| ΔGtot | 50.5 ± 0.8 | 47.2 ± 0.1 |
| ΔGele | 47.0 ± 0.7 | 43.9 ± 0.4 |
| ΔGvdW | 0.5 ± 0.1 | 0.5 ± 0.3 |
| ΔGpmf | 3.0 ± 0.1 | 2.9 ± 0.1 |
| (C) | ΔG(4)cycle3 | ΔG(3)cycle3 |
| CysMeTANDEM:I[mC]I[mC]→CysMeTANDEM:ICIC | I[mC]I[mC]→ICIC | |
| ΔGtot | 348.3 ± 0.9 | 349.0 ± 0.1 |
| ΔGele | 340.3 ± 0.4 | 340.3 ± 0.0 |
| ΔGvdW | 1.3 ± 0.6 | 2.1 ± 0.1 |
| ΔGpmf | 6.7 ± 0.0 | 6.5 ± 0.0 |
The differences in binding free energy, ΔΔGbinding (Equation 2), for the two DNA bis-intercalating agents, 2QN and CysMeTANDEM, are shown in Table 3. The calculated ΔΔGbinding values are in very good qualitative agreement with the experimental data. Thus, binding of 2QN to d(GCGGCGCCGC)2 is disfavored over binding to d(GCGDTDTCGC)2 by 1.8 kcal mol−1, which is in accordance with the ∼100-fold increase in affinity for 2QN binding to a DTDT site compared with a GCGC site in DNA (10). Similarly, binding of CysMeTANDEM to d(CTCICICCAG)2 appears to be even more markedly disfavored (by 4.4 kcal mol−1) over binding to d(CTCATATCAG)2, again in very good agreement with the fact that this ligand binds with very high affinity to TpA steps but does not significantly bind to CpI steps (16).
Table 3.
Averaged differences in binding free energy (kcal mol−1) for 2QN and CysMeTANDEM
| ΔG(4) − ΔG(3) | ΔG(4) − ΔG(3) | ΔG(4) − ΔG(3) | ΔΔGbinding | |
|---|---|---|---|---|
| Cycle 1 | Cycle 2 | Cycle 3 | ||
| 2QN | ||||
| ΔΔGtot | 1.2 | 1.7 | −1.1 | 1.8 |
| ΔΔGele | 0.9 | 2.2 | 0.5 | 3.6 |
| ΔΔGvdW | −0.1 | −0.2 | −1.4 | −1.7 |
| ΔΔGpmf | 0.4 | −0.3 | −0.2 | −0.1 |
| CysMeTANDEM | ||||
| ΔΔGtot | 1.8 | 3.3 | −0.7 | 4.4 |
| ΔΔGele | 0.6 | 3.2 | 0.3 | 4.1 |
| ΔΔGvdW | −0.5 | −0.3 | −0.8 | −1.6 |
| ΔΔGpmf | 1.6 | 0.1 | 0.2 | 1.9 |
Most of the change in free energy for binding of 2QN to d(GCGGCGCCGC)2 and d(GCGDTDTCGC)2 originates from differences in the electrostatic interactions (ΔΔGele, Table 3) calculated for the DTDT→D[mC]GT perturbation (0.9 kcal mol−1) and from conversion of the latter state to G[mC]G[mC] (2.2 kcal mol−1). These unfavorable changes are counterbalanced by changes in the van der Waals term (ΔΔGvdW) mainly affecting the G[mC]G[mC]→GCGC mutation (−1.4 kcal mol−1).
Because the terms ΔΔGvdW and ΔΔGpmf virtually compensate for each other in the binding of CysMeTANDEM to d(CTCATATCAG)2 and d(CTCICICCAG)2 (Table 3), the results clearly point to ΔΔGele as the energy component that is responsible for the notable loss of affinity of CysMeTANDEM toward the ICIC sequence relative to ATAT. Furthermore, the largest contribution to the destabilizing electrostatic term (3.2 kcal mol−1) comes from the charge redistribution that takes place when the distal hypoxanthine:5-methylcytosine pairs are mutated to adenine:thymine (cycle 2). In contrast, the similar mutation affecting the central base pairs (cycle 1) brings about an overall unfavorable free energy change of only 1.8 kcal mol−1, of which just about one-third corresponds to ΔΔGele. It is striking that the ΔGpmf term does not cancel in the CysMeTANDEM cycle as it does in the 2QN cycle but this may be due to differences in the stacking geometries of the central bases as a consequence of the presence of the intercalated chromophores.
Rationale for the sequence preferences
Stacking interactions
The preceding results demonstrate that the MD-TI simulations correctly predict the preferential binding of 2QN to DTDT over GCGC and the much higher affinity of CysMeTANDEM for ATAT relative to ICIC. Furthermore, our calculations point to the electrostatic contribution (ΔΔGele) as the critical determinant of binding selectivity for these two bis-intercalating ligands. In addition, the fact that the highest ΔΔGele value corresponds to the change in the nature of the distal bases (cycle 2) clearly suggests that stacking interactions are crucial in the modulation of the observed binding affinities. Thus, the recognition site for these bis-intercalators definitely extends beyond the central dinucleotide step to cover four base pairs.
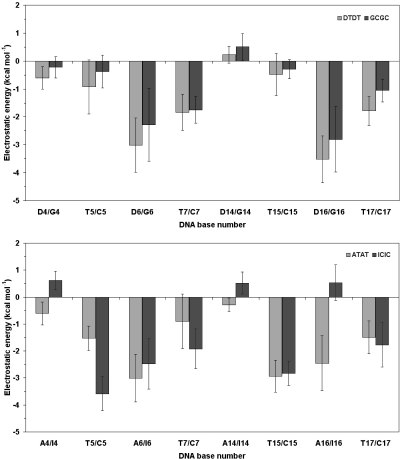
Because the differences in binding free energy between the natural and modified oligonucleotides appeared to arise mostly from variations in the electrostatic component of the stacking interactions between the heteroaromatic rings, we used classical continuum electrostatic theory to examine the influence of solvent-screened electrostatic forces into the association process. Consistent with the MD-TI results, the electrostatic contributions calculated between each bis-intercalating ligand and the individual bases that make up the intercalation sites (Figure 5) are more favorable for DTDT relative to GCGC and also for ATAT over ICIC. However, given the standard errors of the mean values, it is not feasible to discern whether these differences are more notable for the distal base pairs (D4:T17/G4:C17 and T7:D14/C7:G14) or for the central ones (T5:D16/C5:G16 and D6:T15/G6:C15). Nonetheless, the most apparent differences are seen for the interaction of CysMeTANDEM with I4, I14 and I16, which turn out to be noticeably unfavorable (i.e. >0), in contrast with the corresponding interactions with the equivalent purines in ATAT (A4, A14 and A16), which are favorable, particularly for the latter base. A more detailed analysis (data not shown) revealed that most of the destabilization is due to the charge redistribution that accompanies reversal of the hydrogen-bonding character of the N1 atom in going from I to A, as depicted in Figure 2.
Figure 5.
Calculated electrostatic interaction energies between the 2QN (top) and CysMeTANDEM (bottom) and the DNA bases that make up the two intercalation sites in each of the two decanucleotides studied. These results are mean values ± standard deviation (kcal mol−1) from snapshots taken every 2.0 ps from the 1 ns equilibration phase of the MD simulations.
Taken together, the calculations point to the electrostatic component of the stacking interactions as the most important term favoring binding of 2QN and CysMeTANDEM to DTDT and ATAT steps, respectively.
Hydrogen bonding interactions and minor groove desolvation
To gain extra physical insight into the factors that modulate the reported binding preferences, we also examined the possibility that the affinities could be modulated by differences in hydrogen-bonding interactions in the ligand:DNA complexes, as suggested (28). However, the pattern of hydrogen bonds between the depsipeptide residues of 2QN and the central base pairs was found to be very similar in the equilibrated complexes with d(GCGGCGCCGC)2 and d(GCGDTDTCGC)2 (Supplementary Table S1), pointing to the simultaneous occurrence of only three hydrogen bonds in both cases, a finding that is in consonance with previous NMR results for echinomycin (13). Therefore, we can assume that hydrogen bonding does not play a significant role in the modulation of the selective binding of 2QN to DTDT versus GCGC.
As regards CysMeTANDEM, the intramolecular hydrogen bonds that prevent the carbonyl oxygens of its alanine residues from recognizing the 2-amino group of guanine in the minor groove are similarly maintained in the complexes with d(CTCATATCAG)2 and d(CTCICICCAG)2 (Supplementary Table S1). On the contrary, the two intermolecular hydrogen bonding interactions with the purine N3 atoms are apparently better maintained for the TpA site than for the CpI site, although in all cases there are relevant deviations from the optimal geometry expected for a hydrogen-bond interaction. These differences can contribute to the ΔGele changes described above for the mutation affecting the central base pairs (cycle 1) but their magnitude is too small to fully account for the strong selectivity differences determined experimentally. In addition, this free energy change is virtually compensated by a van der Waals difference of similar magnitude but opposite sign (Table 3).
The favorable electrostatic interactions within the ligand–DNA complexes (as in any other drug–receptor complex) (43), and particularly the intermolecular hydrogen bonds, must compensate for the usually unfavorable change in solvation energies that accompanies the association of the interacting partners (46). This is another factor that has been pinpointed as a possible contributor to the differences in sequence selectivity for this class of bis-intercalators (30). In the cases studied here, the favorable electrostatic interactions in the minor groove will be counterbalanced by the desolvation of both the polar groups present in the concave part of the depsipeptides and the edges of the bases that are sandwiched by the drugs. On the other hand, the hydrophobic quinoline or quinoxaline rings of the bis-intercalators are inserted into the hydrophobic environment provided by the space comprised between two contiguous base pairs, which is not exposed to the solvent in the free decamers, and this contribution would be expected to be favorable for binding. These terms, which are implicitly included in the thermodynamic cycles used in TI calculations but cannot be dissected out from the remaining contributions, were estimated by means of Poisson–Boltzmann calculations. When the calculated desolvation energy values for both sets of decamers were compared, no statistically significant differences were observed between ATAT and ICIC or between DTDT and GCGC. Therefore, we can assume that these differences either annihilate or contribute negligibly to the differences in binding free energies of the two bis-intercalating ligands.
Unstacking and creation of the intercalation sites
A last factor that can be thought of as contributing to the differences in binding free energies is the relative ease of unstacking of the DNA steps that make up the intercalation sites. This is so because the energy cost involved in their creation is likely to depend on base composition and sequence (6). Again, this factor is implicitly included in the thermodynamic cycles that were used in the TI calculations but cannot be dissected out from the remaining contributions. To get a rough estimate of possible differential effects in the process of unwinding the double helix and increasing the rise between the base pairs to allow drug binding, a restraint force was applied onto each free DNA decamer during an MD simulation so as to bias the trajectories toward the structure found in each respective drug–DNA complex (see Materials and Methods). The progression of the conformational changes involved in creating the bis-intercalation site was followed by measuring the evolution of the rmsd of the atoms that make up the central tetranucleotide where each drug binds. The rmsd values decreased equally gradually and were almost superimposable for each pair of decanucleotides studied (Supplementary Figure S1) suggesting that, at least under these conditions, the magnitude of this contribution is likely to be similar. Apart from a greater ease of unstacking being apparent in the ATAT-relative to the ICIC-containing sequence (slightly lower rmsd values at a given time or shorter times for achieving a given rmsd value), base pair separation took place when equivalent amounts of energy had been pumped into each system, implying that the energetic cost of intercalation site creation is comparable in all cases. In this respect, it is of interest that the total stabilization energies determined ab initio at high levels of theory for stacked G:C/C:G and A:T/T:A dimers in B-DNA crystal structures have indeed been shown to be very similar (47,48).
Final considerations
From the perspective of a putative ligand, double-helical DNA may be viewed as a rather regular exterior array of phosphate charges surrounding an interior lattice of stacked base pairs that are accessible for binding from either the major or the minor groove. A non-specific electrostatic binding mode is expected between the DNA polyanion and positively charged ligands, which can be followed by the subsequent redistribution to higher-affinity sites along the DNA helix upon recognition of a particular arrangement of functional groups in one or both of the grooves. For echinomycin and related ligands, it has been suggested that the lack of a net positive charge could be compensated by the high dipole moment calculated for these molecules, which arises from an asymmetric distribution of positive and negative electrostatic potential regions (10,49). On the other hand, the high degree of structural preorganization in echinomycin-like molecules enables the two planar quinoxaline-2-carboxamide heteroaromatic rings (or quinoline-2-carboxamide in the case of 2QN), which do not have intercalative properties per se, to be held in position by the cyclic depsipeptide backbone so that simultaneous intercalation is possible through the establishment of hydrogen bonding interactions between the alanine residues of the depsipeptides and the bases in the minor groove (8). Definite proof for this binding mode has been repeatedly provided by X-ray and NMR studies on complexes between short DNA oligonucleotides and echinomycin, triostin A (11), TANDEM and CysMeTANDEM (14,16,50). Nonetheless, the precise thermodynamics of the interaction of these compounds with oligonucleotides of defined sequence remains elusive due to the low solubility of these compounds in aqueous solution (8,27).
On the other hand, ligand–DNA interaction energies are commonly calculated using molecular mechanics either on a single structure that is taken to represent the ensemble average of each complex or on multiple snapshots periodically extracted along an MD trajectory for averaging purposes. Although this latter approach, which is relatively simple, may be useful in detecting trends (31) and can be complemented with continuum methods that consider the desolvation effects that oppose the favorable electrostatic interactions (49), it neglects entropic contributions and does not usually take into account the changes in internal energy undergone by either the drug or the DNA molecules upon binding. For the ligands, these changes are generally assumed to be small and of similar magnitude for all the complexes considered but the situation can be rather different for the DNA molecules due to sequence-dependent microheterogeneity. Thus, intercalation at certain sequences might be favored simply because of the inherent tendency of some dinucleotide steps to underwind and roll (6). Nevertheless, an accurate computation of the binding free energy is hampered by uncertainties regarding their particular conformation in the unbound state, limitations with respect to some energy contributions that are not easily amenable to calculation (e.g. hydrophobic and entropic effects), and the validity of the energy partitioning schemes.
Free energy perturbation and TI allow the most accurate calculations of relative binding strengths and can expose unexpected cancellations in the factors contributing to complex formation (41,51). In contrast with simulations of ligand binding to proteins, which generally need to address substantial molecular reorganization that can span long time scales, the conformational changes involved in the alchemical changes studied here are relatively minor, as they do not significantly affect the double-helical structure of the DNA. In this regard, DNA can be seen as a very well-characterized macromolecular target that provides ample opportunities for the study of stacking interactions using intercalating ligands as probes.
The present work supports earlier suggestions (6,10,31) that the electrostatic contributions to stacking energies can account for the distinct specificity patterns that come to light when bis-intercalating ligands of the type studied here are challenged to bind to DNA molecules possessing identical functionalities in their minor grooves, such as DTDT versus GCGC and ATAT versus ICIC. In addition, it highlights the crucial importance of considering the nature of the flanking bases in order to account for selectivity differences that do not rely on formation of specific hydrogen bonds. In this respect, it must be remarked that the stacking overlap of the quinoline or quinoxaline groups is always substantial relative to the purine bases external to the bis-intercalation sites but much smaller relative to the internal bases, even when the attached carboxamide group is included (12). The rationale exposed here improves our understanding of the stacking interactions involved in the process of DNA sequence recognition by this interesting class of bis-intercalating agents.
CONCLUSIONS
Incorporation of explicit solvent molecules into dynamic models and use of the TI method coupled to MD simulations has made it possible to obtain accurate free energy differences for the binding of 2QN, a quinoline analog of echinomycin, and CysMeTANDEM, a partially demethylated analog of triostin A, to different DNA duplexes. The four oligonucleotides studied incorporate not only standard DNA bases but also hypoxanthine (structurally equivalent to a guanine base that has lost its 2-amino group) and 2,6-diaminopurine (representing an adenine base with an added 2-amino group). The alchemical changes studied involved whole base pairs, and the calculations were performed in a way that allowed the dissection of contributions from each side of the intercalated heteroaromatic rings.
The calculated differences in affinity for 2QN binding to either d(GCGGCGCCGC)2 or d(GCGDTDTCGC)2, as well as for CysMeTANDEM binding to either d(CTCICICCAG)2 or d(CTCATATCAG)2, are in agreement with available experimental data from different sources. The consistent picture obtained for both 2QN and CysMeTANDEM is that the electrostatic component of the stacking interactions between the DNA base pairs and the sandwiched planar moieties of the drugs is heavily modulating the binding affinities detected experimentally. Thus, when 2QN (or a related natural quinoxaline antibiotic) is challenged with DNA steps having identical hydrogen-bond donating groups in the minor groove, preferential binding is observed at tetranucleotide sites that provide optimal stacking properties (e.g. the DTDT sequence). A similar recognition mechanism modulates the sequence selectivity of CysMeTANDEM. This ligand, which does not seek a hydrogen bond donor in the minor groove because the carbonyl groups of its two alanine residues are involved in intramolecular hydrogen bonds with the NHs of the neighboring valines, naturally avoids CpG steps and binds with high affinity to TpA-containing sequences. However, it also steers clear of CpI steps that provide a minor groove environment similar to that of TpA steps. In light of the findings reported here, this sequence discrimination mostly stems from subtle differences in the stacking geometries of the quinoxaline rings of this drug with respect to the neighboring bases due to the lack of the 2-amino-mediated hydrogen bonds. Since the unfavorable stacking interaction with C:I base pairs is not overridden in this case by good intermolecular hydrogen bonds involving the depsipeptide, binding to CpI-containing sequences is strongly disfavored.
We can conclude that the hydrogen bonds between the depsipeptide and the functional groups present in the DNA minor groove need to be correct because there is a penalty when they are missing or wrong, hence their importance in dictating ligand selectivity, but their net contribution to the binding free energy can be easily over-emphasized if the opposing desolvation of the interacting partners in the aqueous environment of a biological system is not taken into account (46) and other factors are neglected. Hydrophobic interactions, on the other hand, clearly promote binding affinity by displacing water molecules that are ordered on the hydrophobic surfaces of both target and ligand into the bulk solvent but the resulting favorable entropic contribution is likely to be similar for all the sequences studied.
The present results support previous claims that the intercalating chromophores of bis-intercalators (or intercalators in general) should be viewed not as simple hydrophobic plates that become sandwiched indiscriminately between the DNA base pairs but as molecular fragments capable of recognizing the distinct electrostatic properties of the paired nucleobases that make up both sides of the intercalation sites (6). In fact, the process of DNA bis-intercalation is ideally suited for the study of stacking interactions as both the intercalating moiety of these ligands and the DNA base pairs are fixed in a definite orientation by the constraints imposed either by the depsipeptide or the sugar–phosphate backbone, respectively. This means that selectivity patterns need to be explained by considering not only the central dinucleotide step that is sandwiched by the intercalating moieties but also the nature of the flanking bases. Thus, in light of the present findings, the reported increases in affinity for echinomycin binding to standard DNA (KD = 5 µM), I + DAP–DNA (KD = 0.7 µM) and DAP–DNA (KD = 0.07 µM) might be interpreted not only on the basis of relative CpG, TpD and CpD content but also as a function of base composition on the outer side of the intercalated quinoxaline ring (i.e. flanking A:T, G:C, D:T and I:C base pairs) (27).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This study is dedicated to Michael Waring for his outstanding lifelong contributions in the field of DNA–ligand interactions. Support from the Spanish CICYT (SAF2003-7219-C02) and the National Foundation for Cancer Research (to F.G.) is gratefully acknowledged. E.M. enjoys a research fellowship from Comunidad de Castilla-La Mancha. The authors thank the University of Alcalá Computing Center and the CIEMAT (Madrid) for generous allowances of computer time on their SGI servers. Funding to pay the Open Access publication charges for this article was provided by Comisión Interministerial de Ciencia y Tecnología.
Conflict of interest statement. None declared.
REFERENCES
- 1.Saenger W. Principles of Nucleic Acid Structure. NY: Springer- Verlag; 1984. [Google Scholar]
- 2.Goodsell D.S. Sequence recognition of DNA by lexitropsins. Curr. Med. Chem. 2001;8:509–516. doi: 10.2174/0929867003373319. [DOI] [PubMed] [Google Scholar]
- 3.Yang X.L., Wang A.H. Structural studies of atom-specific anticancer drugs acting on DNA. Pharmacol. Ther. 1999;83:181–215. doi: 10.1016/s0163-7258(99)00020-0. [DOI] [PubMed] [Google Scholar]
- 4.Ulyanov N.B., James T.L. Statistical analysis of DNA duplex structural features. Methods Enzymol. 1995;261:90–120. doi: 10.1016/s0076-6879(95)61006-5. [DOI] [PubMed] [Google Scholar]
- 5.Young M.A., Ravishanker G., Beveridge D.L., Berman H.M. Analysis of local helix bending in crystal structures of DNA oligonucleotides and DNA-protein complexes. Biophys. J. 1995;68:2454–2468. doi: 10.1016/S0006-3495(95)80427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gago F. Stacking interactions and intercalative DNA binding. Methods. 1998;14:277–292. doi: 10.1006/meth.1998.0584. [DOI] [PubMed] [Google Scholar]
- 7.Braña M.F., Cacho M., Gradillas A., de Pascual-Teresa B., Ramos A. Intercalators as anticancer drugs. Curr. Pharm. Des. 2001;7:1745–1780. doi: 10.2174/1381612013397113. [DOI] [PubMed] [Google Scholar]
- 8.Waring M.J. Molecular Basis of Specificity in Nucleic Acid–Drug Interactions. Dordrecht, The Netherlands: Kluwer Academic; 1990. [Google Scholar]
- 9.Wemmer D.E. Designed sequence-specific minor groove ligands. Annu. Rev. Biophys. Biomol. Struct. 2000;29:439–461. doi: 10.1146/annurev.biophys.29.1.439. [DOI] [PubMed] [Google Scholar]
- 10.Bailly C., Echepare S., Gago F., Waring M.J. Recognition elements that determine affinity and sequence-specific binding to DNA of 2QN, a biosynthetic bis-quinoline analogue of echinomycin. Anticancer Drug Des. 1999;14:291–303. [PubMed] [Google Scholar]
- 11.Wang A.H., Ughetto G., Quigley G.J., Hakoshima T., van der Marel G.A., van Boom J.H., Rich A. The molecular structure of a DNA-triostin A complex. Science. 1984;225:1115–1121. doi: 10.1126/science.6474168. [DOI] [PubMed] [Google Scholar]
- 12.Cuesta-Seijo J.A., Sheldrick G.M. Structures of complexes between echinomycin and duplex DNA. Acta Crystallogr. D Biol. Crystallogr. 2005;61:442–448. doi: 10.1107/S090744490500137X. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert D.E., Feigon J. The DNA sequence at echinomycin binding sites determines the structural changes induced by drug binding: NMR studies of echinomycin binding to [d(ACGTACGT)]2 and [d(TCGATCGA)]2. Biochemistry. 1991;30:2483–2494. doi: 10.1021/bi00223a027. [DOI] [PubMed] [Google Scholar]
- 14.Addess K.J., Sinsheimer J.S., Feigon J. Solution structure of a complex between [N-MeCys3,N-MeCys7]TANDEM and [d(GATATC)]2. Biochemistry. 1993;32:2498–2508. doi: 10.1021/bi00061a006. [DOI] [PubMed] [Google Scholar]
- 15.Addess K.J., Feigon J. Sequence specificity of quinoxaline antibiotics. 1. Solution structure of a 1:1 complex between triostin A and [d(GACGTC)]2 and comparison with the solution structure of the [N-MeCys3,N-MeCys7]TANDEM-[d(GATATC)]2 complex. Biochemistry. 1994;33:12386–12396. doi: 10.1021/bi00207a005. [DOI] [PubMed] [Google Scholar]
- 16.Addess K.J., Feigon J. Sequence specificity of quinoxaline antibiotics. 2. NMR studies of the binding of [N-MeCys3,N-MeCys7]TANDEM and triostin A to DNA containing a CpI step. Biochemistry. 1994;33:12397–12404. doi: 10.1021/bi00207a006. [DOI] [PubMed] [Google Scholar]
- 17.Viswamitra M.A., Kennard O., Cruse W.B.T., Egert E., Sheldrick G.M., Jones P.G., Waring M.J., Wakelin L.P.G., Olsen R.K. Structure of TANDEM and its implication for bifunctional intercalation into DNA. Nature. 1981;289:817–819. doi: 10.1038/289817a0. [DOI] [PubMed] [Google Scholar]
- 18.Low L.C.M., Olsen R.K., Waring M.J. Sequence preferences in the binding to DNA of triostin A and TANDEM as reported by DNase I footprinting. FEBS Lett. 1984;176:414–420. doi: 10.1016/0014-5793(84)81209-0. [DOI] [PubMed] [Google Scholar]
- 19.Addess K.J., Gilbert D.E., Olsen R.K., Feigon J. Proton NMR studies of [N-MeCys3, N-MeCys7]TANDEM binding to DNA oligonucleotides: sequence-specific binding at the TpA site. Biochemistry. 1992;31:339–350. doi: 10.1021/bi00117a005. [DOI] [PubMed] [Google Scholar]
- 20.Waterloh K., Olsen R.K., Fox K.R. Bifunctional intercalator [N-MeCys3, N-MeCys7]TANDEM binds to the dinucleotide TpA. Biochemistry. 1992;31:6246–6253. doi: 10.1021/bi00142a012. [DOI] [PubMed] [Google Scholar]
- 21.Fletcher M.C., Olsen R.K., Fox K.R. Dissociation of the AT-specific bifunctional intercalator [N-MeCys3, N-MeCys7]TANDEM from TpA sites in DNA. Biochem. J. 1995;306:15–19. doi: 10.1042/bj3060015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavesa M., Fox K.R. Preferred binding sites for [N-MeCys3, N-MeCys7]TANDEM determined using a universal footprinting substrate. Anal. Biochem. 2001;293:246–250. doi: 10.1006/abio.2001.5124. [DOI] [PubMed] [Google Scholar]
- 23.Bailly C., Marchand C., Waring M.J. New binding sites for antitumor antibiotics created by relocating the purine 2-amino group in DNA. J. Am. Chem. Soc. 1993;115:3784–3785. [Google Scholar]
- 24.Jennewein S., Waring M.J. Footprinting of echinomycin and actinomycin D on DNA molecules asymmetrically substituted with inosine and/or 2,6-diaminopurine. Nucleic Acids Res. 1997;25:1502–1510. doi: 10.1093/nar/25.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailly C., Waring M.J. DNA recognition by quinoxaline antibiotics: use of base-modified DNA molecules to investigate determinants of sequence-specific binding of triostin A and TANDEM. Biochem. J. 1998;330:81–87. doi: 10.1042/bj3300081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchand C., Bailly C., McLean M., Moroney S.E., Waring M.J. The 2-amino group of guanine is absolutely required for specific binding of the anti-cancer antibiotic echinomycin to DNA. Nucleic Acids Res. 1992;20:5601–5606. doi: 10.1093/nar/20.21.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng Y.D., Ge H., Wang X., Edwardson J.M., Waring M.J., Fitzgerald W.J., Henderson R.M. Atomic force microscopy study of the structural effects induced by echinomycin binding to DNA. J. Mol. Biol. 2005;345:745–758. doi: 10.1016/j.jmb.2004.10.059. [DOI] [PubMed] [Google Scholar]
- 28.Leng F., Chaires J.B., Waring M.J. Energetics of echinomycin binding to DNA. Nucleic Acids Res. 2003;31:6191–6197. doi: 10.1093/nar/gkg826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alfredson T.V., Maki A.H. Phosphorescence and optically detected magnetic resonance studies of echinomycin–DNA complexes. Biochemistry. 1990;29:9052–9064. doi: 10.1021/bi00490a024. [DOI] [PubMed] [Google Scholar]
- 30.Alfredson T.V., Maki A.H., Waring M.J. Optically detected triplet-state magnetic resonance studies of the DNA complexes of the bisquinoline analogue of echinomycin. Biochemistry. 1991;30:9665–9675. doi: 10.1021/bi00104a014. [DOI] [PubMed] [Google Scholar]
- 31.Gallego J., Luque F.J., Orozco M., Burgos C., Alvarez-Builla J., Rodrigo M.M., Gago F. DNA sequence-specific reading by echinomycin: role of hydrogen bonding and stacking interactions. J. Med. Chem. 1994;37:1602–1609. doi: 10.1021/jm00037a010. [DOI] [PubMed] [Google Scholar]
- 32.Gago F., Richards W.G. Netropsin binding to poly[d(IC)]·poly[d(IC)] and poly[d(GC)]·poly[d(GC)]: a computer simulation. Mol. Pharmacol. 1990;37:341–346. [PubMed] [Google Scholar]
- 33.Bayly C.I., Cieplak P., Cornell W.D., Kollman P.A. A well-behaved electrostatic potential based method using charge restraints for determining atom-centered charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 34.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Zakrzewski V.G., Montgomery J.A., Jr, Stratmann R.E., Burant J.C., et al. Gaussian 98. Pittsburgh, PA: Gaussian, Inc.; 2001. revision A.11.2. [Google Scholar]
- 35.Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. A second-generation force field for the simulation of proteins, nucleic acids and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 36.Arnott S., Hukins D.W. Optimised parameters for A-DNA and B-DNA. Biochem. Biophys. Res. Comm. 1972;47:1504–1509. doi: 10.1016/0006-291X(72)90243-4. [DOI] [PubMed] [Google Scholar]
- 37.Åqvist J. Ion-water interaction potentials derived from free energy perturbation simulations. J. Phys. Chem. 1990;94:8021–8024. [Google Scholar]
- 38.Jorgensen W.L., Chandrasekhar J., Madura J.D. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 39.Darden T.A., York D., Pedersen L.G. Particle mesh Ewald: an N*log(N) method for computing Ewald sums. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 40.Ryckaert J.P., Ciccoti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 41.Kollman P. Free energy calculations: applications to chemical and biochemical phenomena. Chem. Rev. 1993;93:2395–2417. [Google Scholar]
- 42.Pearlman D.A., Kollman P.A. The overlooked bond-stretching contribution in free energy perturbation calculations. J. Chem. Phys. 1991;94:4532–4545. [Google Scholar]
- 43.Pérez C., Ortiz A.R., Pastor M., Gago F. Comparative binding energy analysis of HIV-1 protease inhibitors: incorporation of solvent effects and validation as a powerful tool in receptor-based drug design. J. Med. Chem. 1998;41:836–852. doi: 10.1021/jm970535b. [DOI] [PubMed] [Google Scholar]
- 44.Marco E., Laine W., Tardy C., Lansiaux A., Iwao M., Ishibashi F., Bailly C., Gago F. Molecular determinants of topoisomerase I poisoning by lamellarins: comparison with camptothecin and structure-activity relationships. J. Med. Chem. 2005;48:3796–3807. doi: 10.1021/jm049060w. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez-Barrios F., Gago F. Understanding the basis of resistance in the irksome Lys103Asn HIV-1 reverse transcriptase mutant through targeted molecular dynamics simulations. J. Am. Chem. Soc. 2004;126:15386–15387. doi: 10.1021/ja045409t. [DOI] [PubMed] [Google Scholar]
- 46.Honig B., Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 47.Alhambra C., Luque F.J., Gago F., Orozco M. Ab initio study of stacking interactions in A- and B-DNA. J. Phys. Chem. B. 1997;101:3846–3853. [Google Scholar]
- 48.Dabkowska I., Gonzalez H.V., Jurecka P., Hobza P. Stabilization energies of the hydrogen-bonded and stacked structures of nucleic acid base pairs in the crystal geometries of CG, AT, and AC DNA Steps and in the NMR Geometry of the 5′-d(GCGAAGC)-3′ hairpin: complete basis set calculations at the MP2 and CCSD(T) levels. J. Phys. Chem. A. 2005;109:1131–1136. doi: 10.1021/jp046738a. [DOI] [PubMed] [Google Scholar]
- 49.Gallego J., Ortiz A.R., de Pascual-Teresa B., Gago F. Structure-affinity relationships for the binding of actinomycin D to DNA. J. Comput. Aided Mol. Des. 1997;11:114–128. doi: 10.1023/a:1008018106064. [DOI] [PubMed] [Google Scholar]
- 50.Addess K.J., Feigon J. NMR investigation of Hoogsteen base pairing in quinoxaline antibiotic–DNA complexes: comparison of 2:1 echinomycin, triostin A and [N-MeCys3,N-MeCys7] TANDEM complexes with DNA oligonucleotides. Nucleic Acids Res. 1994;22:5484–5491. doi: 10.1093/nar/22.24.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giudice E., Lavery R. Simulations of nucleic acids and their complexes. Acc. Chem. Res. 2002;35:350–357. doi: 10.1021/ar010023y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.