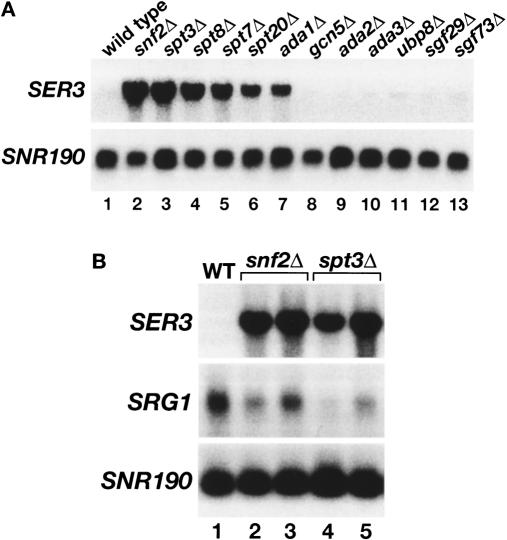
Figure 4.
Repression of SER3 is dependent on the Spt3 and Spt8 subunits of SAGA. (A) Northern analysis of SER3. Total RNA was isolated from wild-type (FY3), snf2Δ (FY1360), spt3Δ (FY294), spt8Δ (FY50), spt7Δ (FY963), spt20Δ (FY1098), ada1Δ (FY1560), gcn5Δ (FY1600), ada2Δ (FY1548), ada3Δ (FY1596), ubp8Δ (FY2473), sgf29Δ (FY2474), and sgf73Δ (FY2475) strains that were grown in YPD. SNR190 served as a loading control. These data are representative of three independent experiments. (B) Northern analysis of SER3 and SRG1. Total RNA was isolated from wild-type (FY4), two snf2Δ (FY2150 and FY2151), and two spt3Δ (FY930 and FY2142) strains that were grown in SD + ser medium. RNA levels were averaged for at least three independent experiments. SER3 mRNA levels are 31.4 ± 2.7 and 27.5 ± 2.3 in snf2Δ and spt3Δ strains, respectively, as compared with wild type. SRG1 RNA levels are 0.59 ± 0.06 and 0.37 ± 0.07 in snf2Δ and spt3Δ strains, respectively, as compared with wild type.