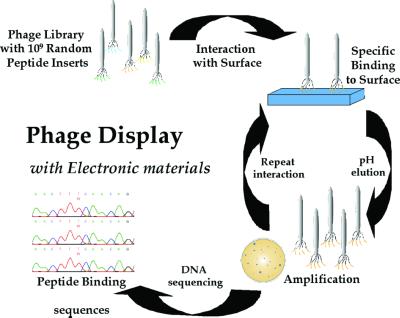
Figure 5.
Peptide selection for electronic materials. The 1.9 × 109 random peptide sequences are exposed to the different crystal substrates; nonspecific peptide interactions are removed with extensive washes. The phage that bind are eluted by lowering the pH and disrupting the surface interaction. The eluted phage are amplified by infecting the E. coli ER2537 host, producing enriched populations of phage, displaying peptides that interact with the specific crystal substrate. The amplified phage are isolated, titered, and reexposed to a freshly prepared substrate surface, thereby enriching the phage population with substrate-specific binding phage. This procedure is repeated three to five times to select the phage with the tightest and most specific binding. The DNA of phage that show specificity is sequenced to determine the peptide-binding sequence.