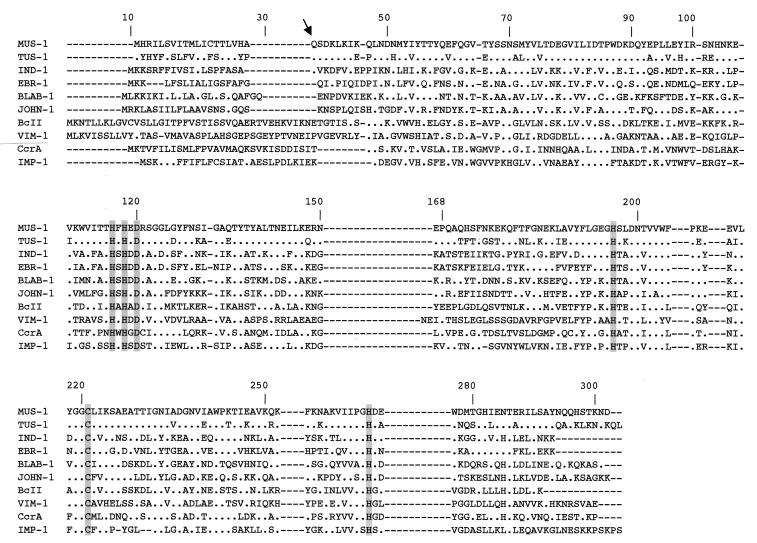
FIG. 1.
Comparison of amino acid sequences of TUS-1 and MUS-1 with those of other metallo-β-lactamases of subclass B1. The origins of the metallo-β-lactamases were as follows: BLAB-1, Chryseobacterium meningosepticum CIP 6058 (32); IND-1, Chryseobacterium indologenes (4); VIM-1, Pseudomonas aeruginosa VT-143/97 (21); BcII, Bacillus cereus 569/H (20); CcrA, Bacteroides fragilis (30); IMP-1, Serratia marcescens TN9106 (27); EBR-1, Empedobacter brevis (GenBank accession no. AF416700); and JOHN-1, Flavobacterium johnsoniae (GenBank accession no. AY028464). The BBL numbering scheme identification numbers are indicated above the sequences (15). The arrow indicates cleavage of the peptide leader site for TUS-1 and MUS-1. Amino acid residues that may be involved in Zn2+ binding appear in grey. Dashes were introduced to optimize the alignment, whereas dots indicate amino acid residues identical to those of β-lactamase MUS-1.